7NVR
 
 | | Human Mediator with RNA Polymerase II Pre-initiation complex | | Descriptor: | CDK-activating kinase assembly factor MAT1, Cyclin-H, Cyclin-dependent kinase 7, ... | | Authors: | Rengachari, S, Schilbach, S, Aibara, S, Cramer, P. | | Deposit date: | 2021-03-15 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVY
 
 | | RNA polymerase II pre-initiation complex with closed promoter DNA in proximal position | | Descriptor: | CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7NW0
 
 | | RNA polymerase II pre-initiation complex with open promoter DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NQH
 
 | | 55S mammalian mitochondrial ribosome with mtRF1a and P-site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
1XSF
 
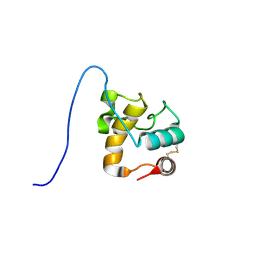 | | Solution structure of a resuscitation promoting factor domain from Mycobacterium tuberculosis | | Descriptor: | Probable resuscitation-promoting factor rpfB | | Authors: | Cohen-Gonsaud, M, Barthe, P, Henderson, B, Ward, J, Roumestand, C, Keep, N.H. | | Deposit date: | 2004-10-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes
Nat.Struct.Mol.Biol., 12, 2005
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
5TXV
 
 | | HslU P21 cell with 4 hexamers | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU | | Authors: | Grant, R.A, Chen, J, Glynn, S.E, Sauer, R.T. | | Deposit date: | 2016-11-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (7.086 Å) | | Cite: | Covalently linked HslU hexamers support a probabilistic mechanism that links ATP hydrolysis to protein unfolding and translocation.
J. Biol. Chem., 292, 2017
|
|
1Y00
 
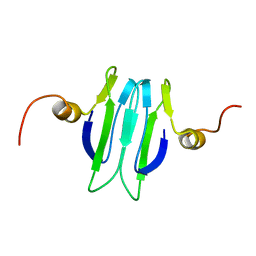 | | Solution structure of the Carbon Storage Regulator protein CsrA | | Descriptor: | Carbon storage regulator | | Authors: | Gutierrez, P, Li, Y, Osborne, M.J, Liu, Q, Gehring, K, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2004-11-13 | | Release date: | 2005-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carbon storage regulator protein CsrA from Escherichia coli.
J.Bacteriol., 187, 2005
|
|
7Y6P
 
 | | Cryo-EM structure if bacterioferritin holoform | | Descriptor: | Bacterioferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Jobichen, C, Sivaraman, J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Bacterioferritin nanocage structures uncover the biomineralization process in ferritins.
Pnas Nexus, 2, 2023
|
|
5UF3
 
 | |
7Y74
 
 | |
5TRG
 
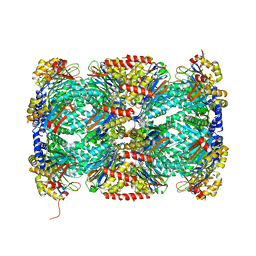 | | Structure of Mycobacterium tuberculosis proteasome in complex with N,C-capped dipeptide DPLG-2 | | Descriptor: | N,N-diethyl-N~2~-[(2E)-3-phenylprop-2-enoyl]-L-asparaginyl-4-fluoro-N-[(naphthalen-1-yl)methyl]-L-phenylalaninamide, Proteasome subunit alpha, Proteasome subunit beta | | Authors: | Hsu, H.-C, Fan, H, Singh, R.K, Wang, R, Sukenick, G, Nathan, C, Lin, G, Li, H. | | Deposit date: | 2016-10-26 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structural Basis for the Species-Selective Binding of N,C-Capped Dipeptides to the Mycobacterium tuberculosis Proteasome.
Biochemistry, 56, 2017
|
|
5W08
 
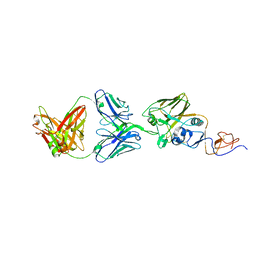 | | A/Texas/50/2012(H3N2) Influenza hemagglutinin in complex with K03.12 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | McCarthy, K.R, Harrison, S.C. | | Deposit date: | 2017-05-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Memory B Cells that Cross-React with Group 1 and Group 2 Influenza A Viruses Are Abundant in Adult Human Repertoires.
Immunity, 48, 2018
|
|
7YC5
 
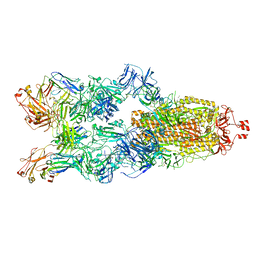 | | Cryo-EM structure of SARS-CoV-2 spike in complex with K202.B bispecific antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain from K202.B, bispecific antibody, ... | | Authors: | Yoo, Y, Cho, H.S. | | Deposit date: | 2022-06-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Novel bispecific human antibody platform specifically targeting a fully open spike conformation potently neutralizes multiple SARS-CoV-2 variants.
Antiviral Res., 212, 2023
|
|
7YBI
 
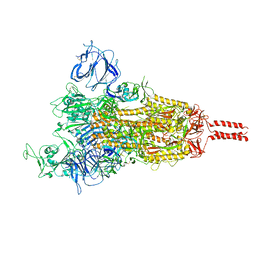 | | SARS-CoV-2 Mu variant spike (open state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, X, Fu, W. | | Deposit date: | 2022-06-29 | | Release date: | 2023-07-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structures of SARS-CoV-2 spike protein alert noteworthy sites for the potential approaching variants.
Virol Sin, 37, 2022
|
|
7NAU
 
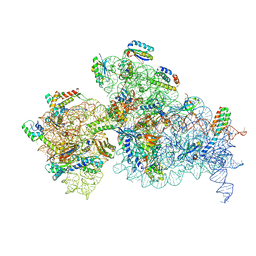 | | Bacterial 30S ribosomal subunit assembly complex state C (Consensus Refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NAV
 
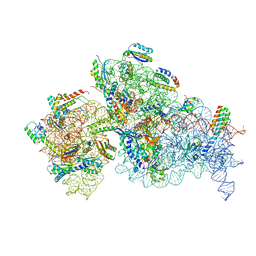 | | Bacterial 30S ribosomal subunit assembly complex state D (Consensus refinement) | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Kaminishi, T, Capuni, R, Astigarraga, E, Gil-Carton, D, Fucini, P, Connell, S. | | Deposit date: | 2021-01-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
7NVZ
 
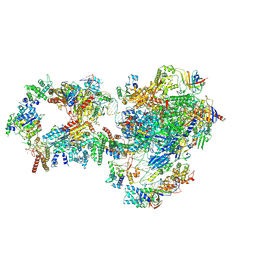 | | RNA polymerase II pre-initiation complex with closed promoter DNA in distal position | | Descriptor: | CDK-activating kinase assembly factor MAT1, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NQL
 
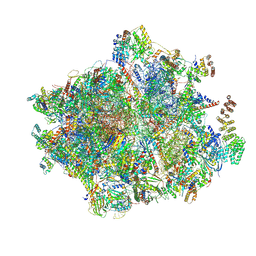 | | 55S mammalian mitochondrial ribosome with ICT1 and P site tRNAMet | | Descriptor: | 12S rRNA, 16S rRNA, 28S ribosomal protein S16, ... | | Authors: | Kummer, E, Schubert, K, Ban, N. | | Deposit date: | 2021-03-01 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of translation termination, rescue, and recycling in mammalian mitochondria.
Mol.Cell, 81, 2021
|
|
7YFI
 
 | | Structure of the Rat tri-heteromeric GluN1-GluN2A-GluN2C NMDA receptor in complex with glycine and glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, M, Zhang, J, Guo, F, Li, Y, Zhu, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Distinct structure and gating mechanism in diverse NMDA receptors with GluN2C and GluN2D subunits.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7OF3
 
 | | Structure of a human mitochondrial ribosome large subunit assembly intermediate in complex with MTERF4-NSUN4 (dataset2). | | Descriptor: | 16S ribosomal RNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Hillen, H.S, Lavdovskaia, E, Nadler, F, Hanitsch, E, Linden, A, Bohnsack, K.E, Urlaub, H, Richter-Dennerlein, R. | | Deposit date: | 2021-05-04 | | Release date: | 2021-05-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of GTPase-mediated mitochondrial ribosome biogenesis and recycling.
Nat Commun, 12, 2021
|
|
7YH7
 
 | | SARS-CoV-2 spike in complex with neutralizing antibody NIV-8 (state 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NIV-8 Fab heavy chain, ... | | Authors: | Moriyama, S, Anraku, Y, Muranishi, S, Adachi, Y, Kuroda, D, Higuchi, Y, Kotaki, R, Tonouchi, K, Yumoto, K, Suzuki, T, Kita, S, Someya, T, Fukuhara, H, Kuroda, Y, Yamamoto, T, Onodera, T, Fukushi, S, Maeda, K, Nakamura-Uchiyama, F, Hashiguchi, T, Hoshino, A, Maenaka, K, Takahashi, Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural delineation and computational design of SARS-CoV-2-neutralizing antibodies against Omicron subvariants.
Nat Commun, 14, 2023
|
|
7Y6T
 
 | | Cryo-EM map of IPEC-J2 cell-derived PEDV PT52 S protein one D0-down and two D0-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hsu, S.T.D, Draczkowski, P, Wang, Y.S. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | In situ structure and dynamics of an alphacoronavirus spike protein by cryo-ET and cryo-EM.
Nat Commun, 13, 2022
|
|
