1CRJ
 
 | |
1CRK
 
 | | MITOCHONDRIAL CREATINE KINASE | | Descriptor: | CREATINE KINASE, PHOSPHATE ION | | Authors: | Fritz-Wolf, K, Schnyder, T, Wallimann, T, Kabsch, W. | | Deposit date: | 1996-03-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of mitochondrial creatine kinase.
Nature, 381, 1996
|
|
1CRL
 
 | |
1CRM
 
 | | STRUCTURE AND FUNCTION OF CARBONIC ANHYDRASES | | Descriptor: | CARBONIC ANHYDRASE I, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Yadava, V.S, Kannan, K.K. | | Deposit date: | 1994-03-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Carbonic Anhydrases
Biomolecular Structure, Conformation, Function and Evolution, 1, 1981
|
|
1CRN
 
 | |
1CRP
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRQ
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRR
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF RAS P21. GDP DETERMINED BY HETERONUCLEAR THREE AND FOUR DIMENSIONAL NMR SPECTROSCOPY | | Descriptor: | C-H-RAS P21 PROTEIN, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Kraulis, P.J, Domaille, P.J, Campbell-Burk, S.L, Van Aken, T, Laue, E.D. | | Deposit date: | 1993-11-24 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1CRU
 
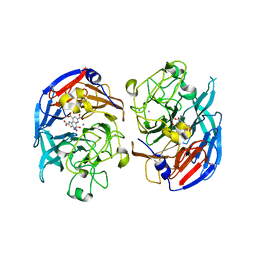 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS IN COMPLEX WITH PQQ AND METHYLHYDRAZINE | | Descriptor: | CALCIUM ION, GLYCEROL, METHYLHYDRAZINE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-16 | | Release date: | 2000-03-01 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CRW
 
 | |
1CRX
 
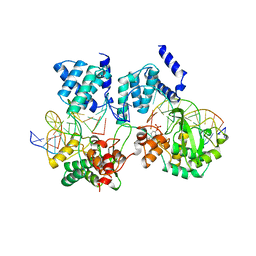 | | CRE RECOMBINASE/DNA COMPLEX REACTION INTERMEDIATE I | | Descriptor: | CRE RECOMBINASE, DNA (5'-D(*AP*TP*AP*TP*GP*CP*TP*AP*TP*AP*CP*GP*AP*AP*GP*TP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*AP*CP*TP*TP*CP*GP*TP*AP*TP*AP*G)-3'), ... | | Authors: | Guo, F, Gopaul, D.N, Van Duyne, G.D. | | Deposit date: | 1997-07-02 | | Release date: | 1998-10-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse.
Nature, 389, 1997
|
|
1CRY
 
 | |
1CRZ
 
 | | CRYSTAL STRUCTURE OF THE E. COLI TOLB PROTEIN | | Descriptor: | TOLB PROTEIN | | Authors: | Abergel, C, Bouveret, E, Claverie, J.-M, Brown, K, Rigal, A, Lazdunski, C, Benedetti, H. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-16 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution.
Structure Fold.Des., 7, 1999
|
|
1CS0
 
 | | Crystal structure of carbamoyl phosphate synthetase complexed at CYS269 in the small subunit with the tetrahedral mimic l-glutamate gamma-semialdehyde | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBAMOYL PHOSPHATE SYNTHETASE: LARGE SUBUNIT, CARBAMOYL PHOSPHATE SYNTHETASE: SMALL SUBUNIT, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 1999-08-16 | | Release date: | 1999-12-10 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The small subunit of carbamoyl phosphate synthetase: snapshots along the reaction pathway.
Biochemistry, 38, 1999
|
|
1CS1
 
 | |
1CS2
 
 | | NMR STRUCTURES OF B-DNA D(CTACTGCTTTAG).D(CTAAAGCAGTAG) | | Descriptor: | 5'-d(*CP*TP*AP*AP*AP*GP*CP*AP*GP*TP*AP*G)-3', 5'-d(*CP*TP*AP*CP*TP*GP*CP*TP*TP*TP*AP*G)-3' | | Authors: | Leporc, S, Mauffret, O, Tevanian, G, Lescot, E, Monnot, M, Fermandjian, S. | | Deposit date: | 1999-08-16 | | Release date: | 1999-08-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR and molecular modelling analysis of d(CTACTGCTTTAG). d(CTAAAGCAGTAG) reveals that the particular behaviour of TpA steps is related to edge-to-edge contacts of their base-pairs in the major groove
Nucleic Acids Res., 27, 1999
|
|
1CS3
 
 | |
1CS4
 
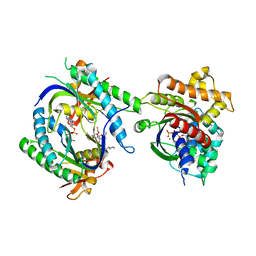 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, PYROPHOSPHATE AND MG | | Descriptor: | 2'-DEOXY-ADENOSINE 3'-MONOPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Dessauer, C.A, Sunahara, R.K, Johnson, R.A, Gilman, A.G, Sprang, S.R. | | Deposit date: | 1999-08-16 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for P-site inhibition of adenylyl cyclase.
Biochemistry, 39, 2000
|
|
1CS6
 
 | | N-TERMINAL FRAGMENT OF AXONIN-1 FROM CHICKEN | | Descriptor: | AXONIN-1, GLYCEROL | | Authors: | Freigang, J, Proba, K, Diederichs, K, Sonderegger, P, Welte, W. | | Deposit date: | 1999-08-17 | | Release date: | 2000-05-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion.
Cell(Cambridge,Mass.), 101, 2000
|
|
1CS7
 
 | | SYNTHETIC DNA HAIRPIN WITH STILBENEDIETHER LINKER | | Descriptor: | 5'-D(GP*(BRU)P*TP*TP*TP*GP*(S02)*CP*AP*AP*AP*AP*C)-3', STRONTIUM ION | | Authors: | Lewis, F.D, Liu, X, Wu, Y, Miller, S.E, Wasielewski, M.R, Letsinger, R.L, Sanishvili, R, Joachimiak, A, Tereshko, V, Egli, M. | | Deposit date: | 1999-08-17 | | Release date: | 2001-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Photoinduced Electron Transfer in Exceptionally Stable Synthetic DNA Hairpins with Stilbenediether Linkers
J.Am.Chem.Soc., 121, 1999
|
|
1CS8
 
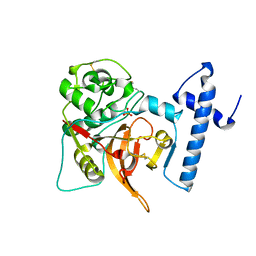 | | CRYSTAL STRUCTURE OF PROCATHEPSIN L | | Descriptor: | HUMAN PROCATHEPSIN L | | Authors: | Cygler, M, Coulombe, R. | | Deposit date: | 1999-08-17 | | Release date: | 1999-08-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment.
EMBO J., 15, 1996
|
|
1CS9
 
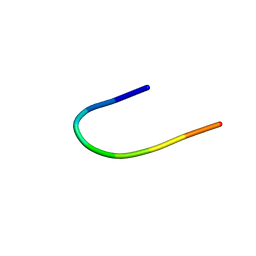 | | SOLUTION STRUCTURE OF CGGIRGERA IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 7 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-18 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CSA
 
 | |
1CSB
 
 | |
1CSC
 
 | |
