1LGX
 
 | | T4 Lysozyme Mutant L99A/M102Q Bound by 3,5-difluoroaniline | | Descriptor: | 3,5-DIFLUOROANILINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Wei, B.Q, Baase, W.A, Weaver, L.H, Matthews, B.W, Shoichet, B.K. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Model Binding Site for Testing Scoring Functions in Molecular Docking
J.Mol.Biol., 322, 2002
|
|
1L19
 
 | |
1LHL
 
 | | ROLE OF PROLINE RESIDUES IN HUMAN LYSOZYME STABILITY: A SCANNING CALORIMETRIC STUDY COMBINED WITH X-RAY STRUCTURE ANALYSIS OF PROLINE MUTANTS | | Descriptor: | HUMAN LYSOZYME | | Authors: | Inaka, K, Matsushima, M, Herning, T, Kuroki, R, Yutani, K, Kikuchi, M. | | Deposit date: | 1992-03-27 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of proline residues in human lysozyme stability: a scanning calorimetric study combined with X-ray structure analysis of proline mutants.
Biochemistry, 31, 1992
|
|
4ND5
 
 | |
1L1Q
 
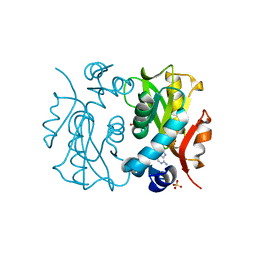 | | Crystal Structure of APRTase from Giardia lamblia Complexed with 9-deazaadenine | | Descriptor: | 9-DEAZAADENINE, Adenine phosphoribosyltransferase, SULFATE ION | | Authors: | Shi, W, Sarver, A.E, Wang, C.C, Tanaka, K.S, Almo, S.C, Schramm, V.L. | | Deposit date: | 2002-02-19 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Closed Site Complexes of Adenine Phosphoribosyltransferase from Giardia lamblia
Reveal a Mechanism of Ribosyl Migration.
J.Biol.Chem., 277, 2002
|
|
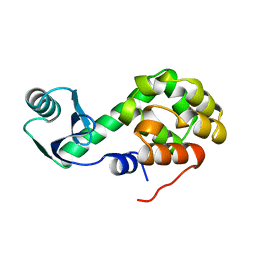
1L22
 
 | |
5N6N
 
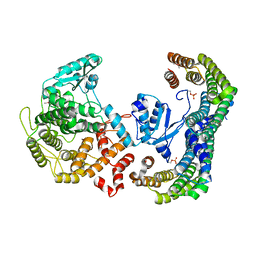 | | CRYSTAL STRUCTURE OF THE 14-3-3:NEUTRAL TREHALASE NTH1 COMPLEX | | Descriptor: | CALCIUM ION, Neutral trehalase, Protein BMH1, ... | | Authors: | Alblova, M, Smidova, A, Obsilova, V, Obsil, T. | | Deposit date: | 2017-02-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Molecular basis of the 14-3-3 protein-dependent activation of yeast neutral trehalase Nth1.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4NKM
 
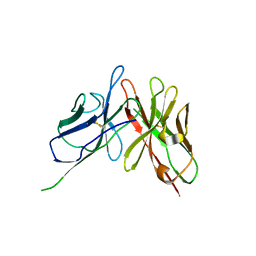 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
1L2C
 
 | | MutM (Fpg)-DNA Estranged Thymine Mismatch Recognition Complex | | Descriptor: | 5'-D(*AP*G*GP*TP*AP*GP*AP*CP*TP*TP*GP*GP*AP*CP*GP*C)-3', 5'-D(*TP*GP*C*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*TP*AP*CP*C)-3', MutM, ... | | Authors: | Fromme, J.C, Verdine, G.L. | | Deposit date: | 2002-02-20 | | Release date: | 2002-06-14 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM.
Nat.Struct.Biol., 9, 2002
|
|
2WIV
 
 | | Cytochrome-P450 XplA heme domain P21 | | Descriptor: | CYTOCHROME P450-LIKE PROTEIN XPLA, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sabbadin, F, Jackson, R, Bruce, N.C, Grogan, G. | | Deposit date: | 2009-05-18 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.5-A Structure of Xpla-Heme, an Unusual Cytochrome P450 Heme Domain that Catalyzes Reductive Biotransformation of Royal Demolition Explosive.
J.Biol.Chem., 284, 2009
|
|
3UJF
 
 | | Asymmetric complex of human neuron specific enolase-4-PGA/PEP | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Qin, J, Chai, G, Brewer, J, Lovelace, L, Lebioda, L. | | Deposit date: | 2011-11-07 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of asymmetric complexes of human neuron specific enolase with resolved substrate and product and an analogous complex with two inhibitors indicate subunit interaction and inhibitor cooperativity.
J.Inorg.Biochem., 111, 2012
|
|
2WJ1
 
 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(4-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(4-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
4NFY
 
 | |
4NGZ
 
 | | Previously de-ionized HEW lysozyme crystallized in 0.5 M YbCl3/30% (v/v) glycerol and collected at 125K | | Descriptor: | CHLORIDE ION, Lysozyme C, YTTERBIUM (III) ION | | Authors: | Benas, P, Legrand, L, Ries-Kautt, M. | | Deposit date: | 2013-11-03 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Weak protein-cationic co-ion interactions addressed by X-ray crystallography and mass spectrometry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4A5Q
 
 | | Crystal structure of the chitinase Chi1 fitted into the 3D structure of the Yersinia entomophaga toxin complex | | Descriptor: | CHI1 | | Authors: | Busby, J.N, Landsberg, M.J, Simpson, R.M, Jones, S.A, Hankamer, B, Hurst, M.R.H, Lott, J.S. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-16 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Structural Analysis of Chi1 Chitinase from Yen-Tc: The Multisubunit Insecticidal Abc Toxin Complex of Yersinia Entomophaga.
J.Mol.Biol., 415, 2012
|
|
3UR5
 
 | | Crystal Structure of PTE mutant K185R/I274N | | Descriptor: | COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
5N1P
 
 | | Crystal structure of the polysaccharide deacetylase Bc1974 from Bacillus cereus in complex with N-hydroxynaphthalene-1-carboxamide | | Descriptor: | 1,2-ETHANEDIOL, Peptidoglycan N-acetylglucosamine deacetylase, SODIUM ION, ... | | Authors: | Giastas, P, Andreou, A, Eliopoulos, E.E. | | Deposit date: | 2017-02-06 | | Release date: | 2018-02-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.448 Å) | | Cite: | Structures of the Peptidoglycan N-Acetylglucosamine Deacetylase Bc1974 and Its Complexes with Zinc Metalloenzyme Inhibitors.
Biochemistry, 57, 2018
|
|
4A7P
 
 | | Se-Met derivatized UgdG, UDP-glucose dehydrogenase from Sphingomonas elodea | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE DEHYDROGENASE | | Authors: | Rocha, J, Granja, A.T, Sa-Correia, I, Fialho, A.M, Frazao, C. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-21 | | Last modified: | 2019-09-25 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | The Structure of Ugdg, an Udp-Glucose Dehydrogenase from Sphingomonas Elodea Atcc 31461
To be Published
|
|
3URQ
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V/R67H with cyclohexyl methylphosphonate inhibitor | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
1L20
 
 | |
1L04
 
 | |
3US8
 
 | | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | Isocitrate dehydrogenase [NADP], SULFATE ION | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-23 | | Release date: | 2011-12-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of an isocitrate dehydrogenase from Sinorhizobium meliloti 1021
To be Published
|
|
3USS
 
 | |
1L29
 
 | |
4NKD
 
 | | Crystal structure of engineered anti-EE scFv antibody fragment | | Descriptor: | Engineered scFv | | Authors: | Kalyoncu, S, Hyun, J, Pai, J.C, Johnson, J.L, Etzminger, K, Jain, A, Heaner Jr, D, Molares, I.A, Truskett, T.M, Maynard, J.A, Lieberman, R.L. | | Deposit date: | 2013-11-12 | | Release date: | 2014-03-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.303 Å) | | Cite: | Effects of protein engineering and rational mutagenesis on crystal lattice of single chain antibody fragments.
Proteins, 82, 2014
|
|
