7WRC
 
 | | Mouse TRPM8 in LMNG in the presence of calcium, icilin and PI(4,5)P2 | | Descriptor: | CALCIUM ION, Icilin, Transient receptor potential cation channel subfamily M member 8 | | Authors: | Zhao, C, Xie, Y, Guo, J. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Structures of a mammalian TRPM8 in closed state.
Nat Commun, 13, 2022
|
|
7WRE
 
 | |
6HWT
 
 | | Crystal structure of p38alpha in complex with a reduced photoswitchable 2-Azothiazol-based Inhibitor (compound 31) | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-(2-phenylhydrazinyl)-1,3-thiazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Mueller, M.P, Rauh, D. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Azo-, 2-diazocine-thiazols and 2-azo-imidazoles as photoswitchable kinase inhibitors: limitations and pitfalls of the photoswitchable inhibitor approach.
Photochem. Photobiol. Sci., 18, 2019
|
|
1Q6U
 
 | | Crystal structure of FkpA from Escherichia coli | | Descriptor: | CESIUM ION, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-14 | | Release date: | 2004-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
3RJE
 
 | | Ternary complex of DNA Polymerase Beta with a gapped DNA containing 8odG at template position | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*(8OG)P*TP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*A)-3'), DNA (5'-D(P*GP*TP*CP*GP*G)-3'), ... | | Authors: | Batra, V.K, Beard, W.A, Wilson, S.H. | | Deposit date: | 2011-04-15 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1Q6I
 
 | | Crystal structure of a truncated form of FkpA from Escherichia coli, in complex with immunosuppressant FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FKBP-type peptidyl-prolyl cis-trans isomerase fkpA | | Authors: | Saul, F.A, Arie, J.-P, Vulliez-le Normand, B, Kahn, R, Betton, J.-M, Bentley, G.A. | | Deposit date: | 2003-08-13 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional studies of FkpA from Escherichia coli, a cis/trans peptidyl-prolyl isomerase with chaperone activity.
J.Mol.Biol., 335, 2004
|
|
3R16
 
 | | Human CAII bound to N-(4-sulfamoylphenyl)-2-(thiophen-2-yl) acetamide | | Descriptor: | Carbonic anhydrase 2, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Biswas, S, McKenna, R, Supuran, C.T. | | Deposit date: | 2011-03-09 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformational variability of different sulfonamide inhibitors with thienyl-acetamido moieties attributes to differential binding in the active site of cytosolic human carbonic anhydrase isoforms.
Bioorg.Med.Chem., 19, 2011
|
|
3R5S
 
 | | Crystal structure of apo-ViuP | | Descriptor: | Ferric vibriobactin ABC transporter, periplasmic ferric vibriobactin-binding protein | | Authors: | Li, N, Zhang, C, Li, B, Liu, X, Huang, Y, Xu, S, Gu, L. | | Deposit date: | 2011-03-19 | | Release date: | 2012-02-08 | | Last modified: | 2012-05-30 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response.
J.Biol.Chem., 287, 2012
|
|
8SA2
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 1 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 1 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA4
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 3 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 3 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA3
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 2 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 2 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA6
 
 | | apo form of adenosylcobalamin riboswitch dimer | | Descriptor: | apo form of adenosylcobalamin riboswitch dimer | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (5.3 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
8SA5
 
 | | Adenosylcobalamin-bound riboswitch dimer, form 4 | | Descriptor: | Adenosylcobalamin, adenosylcobalamin riboswitch form 4 | | Authors: | Ding, J, Deme, J.C, Stagno, J.R, Yu, P, Lea, S.M, Wang, Y.X. | | Deposit date: | 2023-03-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Capturing heterogeneous conformers of cobalamin riboswitch by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
5XC0
 
 | |
6IJP
 
 | | The structure of the ADAL-IMP complex | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Xie, W, Jia, Q. | | Deposit date: | 2018-10-11 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Alternative conformation induced by substrate binding for Arabidopsis thalianaN6-methyl-AMP deaminase.
Nucleic Acids Res., 47, 2019
|
|
2XUT
 
 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
1QWU
 
 | | Golgi alpha-mannosidase II D341N mutant complex with 5-F-guloside | | Descriptor: | (2R,3S,4R,5S)-2,6-difluoro-2-(hydroxymethyl)oxane-3,4,5-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Numao, S, Kuntz, D.A, Withers, S.G, Rose, D.R. | | Deposit date: | 2003-09-03 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural analysis of covalent reaction intermediates.
J.Biol.Chem., 278, 2003
|
|
3RY1
 
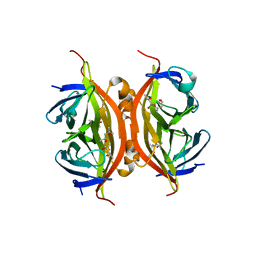 | | Wild-type core streptavidin at atomic resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | Stenkamp, R.E, Wang, Z, Le Trong, I, Stayton, P.S, Lybrand, T.P. | | Deposit date: | 2011-05-10 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Streptavidin and its biotin complex at atomic resolution.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
7YRS
 
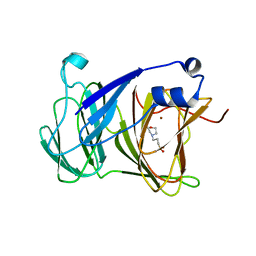 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
1QV4
 
 | |
7Z3B
 
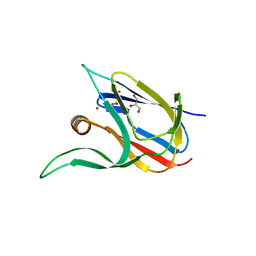 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, reduced form | | Descriptor: | ACETATE ION, AcoP, COPPER (I) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3F
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, oxidized form | | Descriptor: | ACETATE ION, AcoP, CHLORIDE ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3G
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, H166A mutant | | Descriptor: | AcoP, COPPER (I) ION, GLYCEROL | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
7Z3I
 
 | | Crystal structure of the cupredoxin AcoP from Acidithiobacillus ferrooxidans, M171A mutant | | Descriptor: | ACETATE ION, AcoP, COPPER (II) ION, ... | | Authors: | Leone, P, Sciara, G, Ilbert, M. | | Deposit date: | 2022-03-02 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Beyond the coupled distortion model: structural analysis of the single domain cupredoxin AcoP, a green mononuclear copper centre with original features.
Dalton Trans, 53, 2024
|
|
1QV8
 
 | | B-DNA Dodecamer d(CGCGAATTCGCG)2 complexed with proamine | | Descriptor: | 2'-(4-DIMETHYLAMINOPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3' | | Authors: | Squire, C.J, Clark, G.R. | | Deposit date: | 2003-08-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | In vitro studies with methylproamine: a potent new radioprotector.
Cancer Res., 64, 2004
|
|
