7CZZ
 
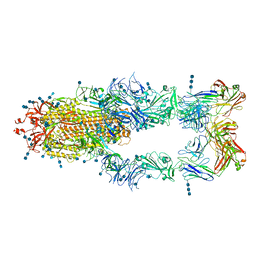 | | S protein of SARS-CoV-2 in complex bound with P5A-2F11_3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
3AB4
 
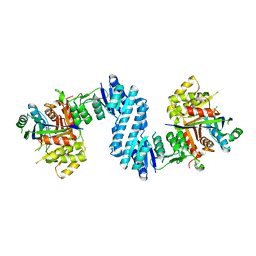 | | Crystal structure of feedback inhibition resistant mutant of aspartate kinase from Corynebacterium glutamicum in complex with lysine and threonine | | Descriptor: | Aspartokinase, LYSINE, THREONINE | | Authors: | Yoshida, A, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-11-30 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mechanism of concerted inhibition of {alpha}2{beta}2-type heterooligomeric aspartate kinase from Corynebacterium glutamicum
J.Biol.Chem., 285, 2010
|
|
3JTL
 
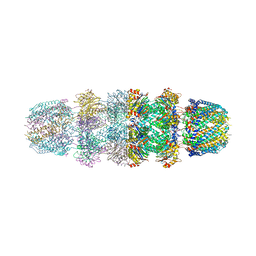 | |
5M92
 
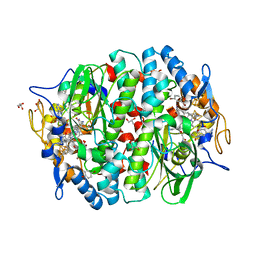 | | PCE reductive dehalogenase from S. multivorans in complex with 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, BENZAMIDINE, BROMIDE ION, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-10-31 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
7D00
 
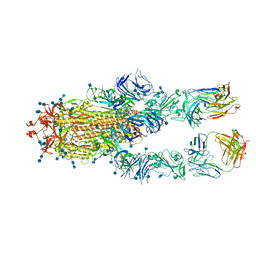 | | S protein of SARS-CoV-2 in complex bound with FabP5A-1B8 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IG c642_heavy_IGHV3-53_IGHD1-26_IGHJ6,chain H of FabP5A-1B8,IGH@ protein, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Zhou, Q. | | Deposit date: | 2020-09-09 | | Release date: | 2021-03-10 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for bivalent binding and inhibition of SARS-CoV-2 infection by human potent neutralizing antibodies.
Cell Res., 31, 2021
|
|
2HEQ
 
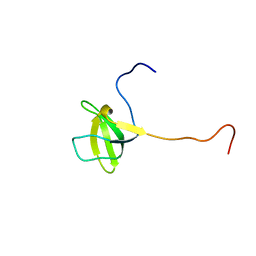 | | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399. | | Descriptor: | YorP protein | | Authors: | Ramelot, T.A, Cort, J.R, Wang, D, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.M, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-21 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of Bacillus subtilis protein YorP, Northeast Structural Genomics Target SR399.
To be Published
|
|
3JVL
 
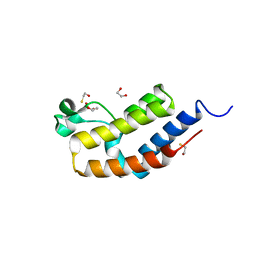 | | Crystal structure of bromodomain 2 of mouse Brd4 | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Bromodomain-containing protein 4 | | Authors: | Vollmuth, F, Blankenfeldt, W, Geyer, M. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structures of the Dual Bromodomains of the P-TEFb-activating Protein Brd4 at Atomic Resolution
J.Biol.Chem., 284, 2009
|
|
5MB3
 
 | |
3ALA
 
 | | Crystal structure of vascular adhesion protein-1 in space group C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ernberg, K.E, McGrath, A.P, Guss, J.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A new crystal form of human vascular adhesion protein 1
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7BB1
 
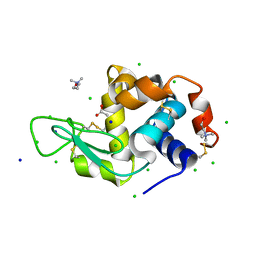 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Glutamic acid 2:1 | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Carrozzini, B. | | Deposit date: | 2020-12-16 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
3ANG
 
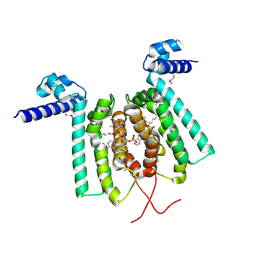 | | Crystal structure of Thermus thermophilus FadR in complex with E. coli-derived dodecyl-CoA | | Descriptor: | DODECYL-COA, Transcriptional repressor, TetR family | | Authors: | Agari, Y, Sakamoto, K, Agari, K, Kuramitsu, S, Shinkai, A. | | Deposit date: | 2010-09-01 | | Release date: | 2011-03-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | TetR-family transcriptional repressor Thermus thermophilus FadR controls fatty acid degradation.
Microbiology, 157, 2011
|
|
7BAZ
 
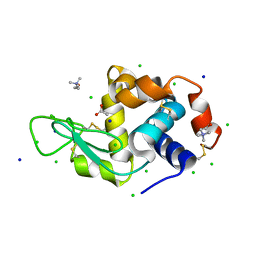 | | Lysozyme crystallized in the presence of the hydrated deep eutectic solvent Choline chloride-Glycerol 1:2 | | Descriptor: | ACETATE ION, CHLORIDE ION, CHOLINE ION, ... | | Authors: | Belviso, B.D, Caliandro, R, Carrozzini, B. | | Deposit date: | 2020-12-16 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Introducing Protein Crystallization in Hydrated Deep Eutectic Solvents
Acs Sustain Chem Eng, 9, 2021
|
|
5MIW
 
 | | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uracil at 1.28 A. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Prokofev, I.I, Lashkov, A.A, Balaev, V.V, Gabdoulkhakov, A.G, Betzel, C, Mikhailov, A.M. | | Deposit date: | 2016-11-29 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | X-ray structure uridine phosphorylase from Vibrio cholerae in complex with uracil at 1.28 A.
To Be Published
|
|
5MK9
 
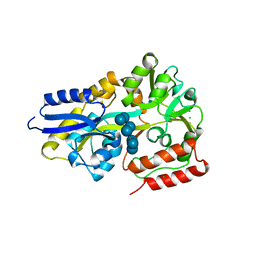 | | Maltodextrin binding protein MalE1 from L. casei BL23 bound to beta-cyclodextrin | | Descriptor: | CHLORIDE ION, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), MalE1 | | Authors: | Homburg, C, Bommer, M, Wuttge, S, Hobe, C, Beck, S, Dobbek, H, Deutscher, J, Licht, A, Schneider, E. | | Deposit date: | 2016-12-02 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | Inducer exclusion in Firmicutes: insights into the regulation of a carbohydrate ATP binding cassette transporter from Lactobacillus casei BL23 by the signal transducing protein P-Ser46-HPr.
Mol. Microbiol., 105, 2017
|
|
3A8K
 
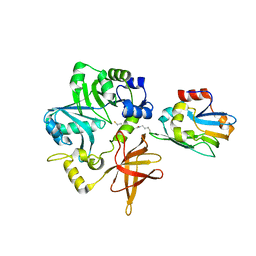 | | Crystal Structure of ETD97N-EHred complex | | Descriptor: | Aminomethyltransferase, Glycine cleavage system H protein | | Authors: | Okamura-Ikeda, K, Hosaka, H. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aminomethyltransferase in complex with dihydrolipoyl-H-protein of the glycine cleavage system: implications for recognition of lipoyl protein substrate, disease-related mutations, and reaction mechanism
J.Biol.Chem., 285, 2010
|
|
3A9Q
 
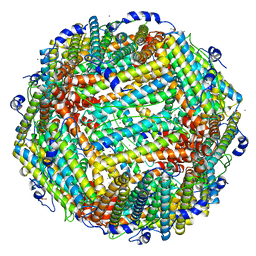 | | Crystal Structure Analysis of E173A variant of the soybean ferritin SFER4 | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-11-05 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
2GSQ
 
 | |
7R5W
 
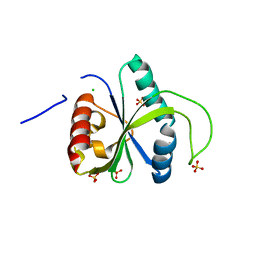 | | Crystal structure of YTHDF2 with compound YLI_DF_029 | | Descriptor: | 6-cyclopropyl-1H-pyrimidine-2,4-dione, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nai, F, Li, Y, Caflisch, A. | | Deposit date: | 2022-02-11 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Fragment Ligands of the m 6 A-RNA Reader YTHDF2.
Acs Med.Chem.Lett., 13, 2022
|
|
7SAS
 
 | | Cryo-EM structure of TMEM106B fibrils extracted from a FTLD-TDP patient, polymorph 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Transmembrane protein 106B | | Authors: | Cao, Q, Jiang, Y, Sawaya, M.R, Eisenberg, D.S. | | Deposit date: | 2021-09-23 | | Release date: | 2022-03-09 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Amyloid fibrils in FTLD-TDP are composed of TMEM106B and not TDP-43.
Nature, 605, 2022
|
|
7BHY
 
 | | DNA-binding domain of DeoR in complex with the DNA operator | | Descriptor: | DNA operator - strand 1, DNA operator - strand 2, Deoxyribonucleoside regulator, ... | | Authors: | Novakova, M, Rezacova, P, Skerlova, J, Brynda, J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into DNA recognition by bacterial transcriptional regulators of the SorC/DeoR family.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
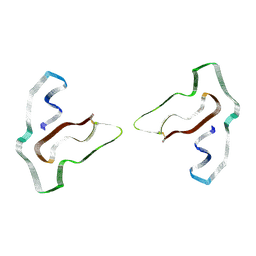
3K3A
 
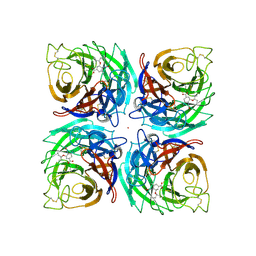 | |
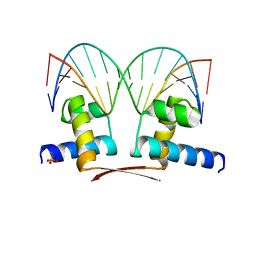
5MJ0
 
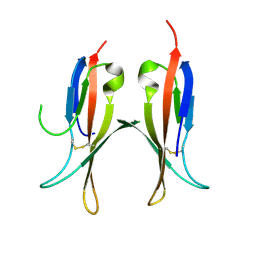 | |
2JGF
 
 | | Crystal structure of mouse acetylcholinesterase inhibited by non-aged fenamiphos | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, HEXAETHYLENE GLYCOL, ... | | Authors: | Hornberg, A, Tunemalm, A.-K, Ekstrom, F. | | Deposit date: | 2007-02-13 | | Release date: | 2007-04-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Acetylcholinesterase in Complex with Organophosphorus Compounds Suggest that the Acyl Pocket Modulates the Aging Reaction by Precluding the Formation of the Trigonal Bipyramidal Transition State.
Biochemistry, 46, 2007
|
|
7LRI
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with DNA, dTTP, and CA(2+) ion | | Descriptor: | CALCIUM ION, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Hoang, A, Ruiz, F.X, Arnold, E. | | Deposit date: | 2021-02-16 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural basis of HIV inhibition by L-nucleosides: Opportunities for drug development and repurposing.
Drug Discov Today, 27, 2022
|
|
2OWN
 
 | |
