2YNA
 
 | |
3K99
 
 | | HSP90 N-terminal domain in complex with 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol | | Descriptor: | 4-(1,3-dihydro-2H-isoindol-2-ylcarbonyl)benzene-1,3-diol, Heat shock protein HSP 90-alpha | | Authors: | Gajiwala, K.S, Davies II, J.F. | | Deposit date: | 2009-10-15 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dihydroxyphenylisoindoline amides as orally bioavailable inhibitors of the heat shock protein 90 (hsp90) molecular chaperone.
J.Med.Chem., 53, 2010
|
|
2NC1
 
 | |
7C07
 
 | | Crystal structure of yeast U2AF1 complex bound to 5'-AAGGU RNA. | | Descriptor: | RNA (5'-R(*U*AP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
5M6Y
 
 | | Cocrystal structure of cAMP-dependent Protein Kinase (PKA) in complex with a methylisoquinoline Fasudil-derivative | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(1,4-diazepan-1-ylsulfonyl)-4-methyl-isoquinoline, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Wienen-Schmidt, B, Heine, A, Klebe, G. | | Deposit date: | 2016-10-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.367 Å) | | Cite: | Cocrystal structure of cAMP-dependent Protein Kinase (PKA) in complex with differently methylated Fasudil-derived ligands
To be Published
|
|
2YN5
 
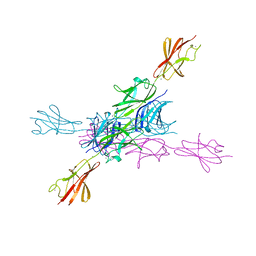 | | Structural insight into the giant calcium-binding adhesin SiiE: implications for the adhesion of Salmonella enterica to polarized epithelial cells | | Descriptor: | CALCIUM ION, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Griessl, M.H, Schmid, B, Kassler, K, Braunsmann, C, Ritter, R, Barlag, B, Sturm, K.U, Danzer, C, Wagner, C, Schaeffer, T.E, Sticht, H, Hensel, M, Muller, Y.A. | | Deposit date: | 2012-10-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insight Into the Giant Ca(2+)-Binding Adhesin Siie: Implications for the Adhesion of Salmonella Enterica to Polarized Epithelial Cells.
Structure, 21, 2013
|
|
5M82
 
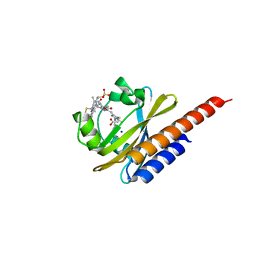 | | Three-dimensional structure of the photoproduct state of GAF3 from Slr1393 of Synechocystis sp. PCC6803 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, PHYCOCYANOBILIN, SODIUM ION, ... | | Authors: | Xu, X.-L, Zhao, K.-H, Gaertner, W, Hoeppner, A. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural elements regulating the photochromicity in a cyanobacteriochrome
Proc.Natl.Acad.Sci.USA, 2020
|
|
2YIR
 
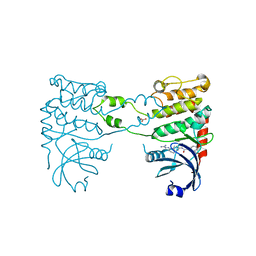 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1352 | | Descriptor: | (E)-N-(5-(2-CARBAMIMIDOYLHYDRAZONO)-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
7QLN
 
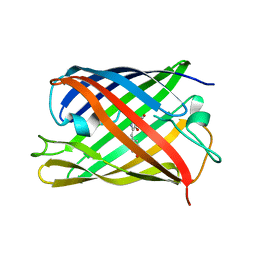 | | rsKiiro pump probe structure by TR-SFX | | Descriptor: | rsKiiro | | Authors: | van Thor, J.J. | | Deposit date: | 2021-12-20 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Optical control of ultrafast structural dynamics in a fluorescent protein.
Nat.Chem., 15, 2023
|
|
2N67
 
 | |
2G7O
 
 | | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM | | Descriptor: | Protein traM | | Authors: | Lu, J, Edwards, R.A, Wong, J.J, Manchak, J, Scott, P.G, Frost, L.S, Glover, J.N. | | Deposit date: | 2006-02-28 | | Release date: | 2006-06-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Protonation-mediated structural flexibility in the F conjugation regulatory protein, TraM.
Embo J., 25, 2006
|
|
5M84
 
 | | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog | | Descriptor: | Eukaryotic translation initiation factor 4E, GLYCEROL, [[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-sulfanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-7-methyl-6-oxidanyl-purin-7-ium-9-yl)-4-methoxy-3-oxidanyl-oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Warminski, M, Nowak, E, Kowalska, J, Jemielity, J, Nowotny, M. | | Deposit date: | 2016-10-28 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Translation initiation factor 4E in complex with (SP)-m2(7,2'O)GppSpA mRNA 5' cap analog
To Be Published
|
|
2YIV
 
 | | NI,FE-CODH with n-butylisocyanate state | | Descriptor: | CARBON MONOXIDE DEHYDROGENASE 2, FE (III) ION, FE(3)-NI(1)-S(4) CLUSTER, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2011-05-16 | | Release date: | 2012-03-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | N-Butyl Isocyanide Oxidation at the [Nife4S4Oh(X)] Cluster of Co Dehydrogenase.
J.Biol.Inorg.Chem., 17, 2012
|
|
5MHM
 
 | |
3JQY
 
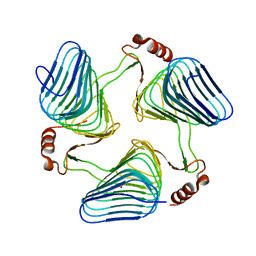 | | Crystal Structure of the polySia specific acetyltransferase NeuO | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Polysialic acid O-acetyltransferase | | Authors: | Schulz, E.-C, Bergfeld, A, Muehlenhoff, M, Ficner, R. | | Deposit date: | 2009-09-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Crystal structure analysis of the polysialic acid specific O-acetyltransferase NeuO
PLoS ONE, 6, 2011
|
|
8QYA
 
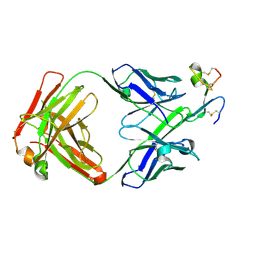 | |
8RI0
 
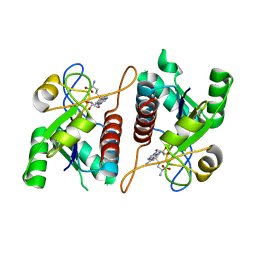 | |
2YBJ
 
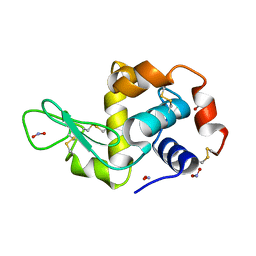 | |
2YBQ
 
 | | The x-ray structure of the SAM-dependent uroporphyrinogen III methyltransferase NirE from Pseudomonas aeruginosa in complex with SAH and uroporphyrinogen III | | Descriptor: | METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE, UROPORPHYRINOGEN III | | Authors: | Storbeck, S, Saha, S, Krausze, J, Klink, B.U, Heinz, D.W, Layer, G. | | Deposit date: | 2011-03-09 | | Release date: | 2011-06-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Heme D1 Biosynthesis Enzyme Nire in Complex with its Substrate Reveals New Insights Into the Catalytic Mechanism of S-Adenosyl-L-Methionine-Dependent Uroporphyrinogen III Methyltransferases.
J.Biol.Chem., 286, 2011
|
|
3JWT
 
 | | Structure of rat neuronal nitric oxide synthase R349A mutant heme domain in complex with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
2YCS
 
 | | Crystal structure of checkpoint kinase 2 in complex with PV788 | | Descriptor: | N-{4-[(1E)-N-CARBAMIMIDOYLETHANEHYDRAZONOYL]PHENYL}-1H-INDOLE-3-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
2NDM
 
 | | NMR solution structure of PawS Derived Peptide 21 (PDP-21) | | Descriptor: | PawS derived peptide 21 | | Authors: | Franke, B, Jayasena, A.S, Fisher, M.F, Swedberg, J.E, Taylor, N.L, Mylne, J.S, Rosengren, K. | | Deposit date: | 2016-07-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Diverse cyclic seed peptides in the Mexican zinnia (Zinnia haageana).
Biopolymers, 106, 2016
|
|
7RB2
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU in BIS-Tris pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-07-05 | | Release date: | 2021-07-14 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 51, 2023
|
|
5MA1
 
 | | PCE reductive dehalogenase from S. multivorans in complex with 2,4,6-trichlorophenol | | Descriptor: | 2,4,6-trichlorophenol, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-11-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
3JX1
 
 | | Structure of rat neuronal nitric oxide synthase D597N mutant heme domain in complex with N1-{(3'R,4'R)-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(3'-fluorophenethyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-{(3R,4R)-4-[(6-amino-4-methylpyridin-2-yl)methyl]pyrrolidin-3-yl}-N'-[2-(3-fluorophenyl)ethyl]ethane-1,2-diamine, ... | | Authors: | Delker, S.L, Li, H, Poulos, T.L. | | Deposit date: | 2009-09-18 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected binding modes of nitric oxide synthase inhibitors effective in the prevention of a cerebral palsy phenotype in an animal model.
J.Am.Chem.Soc., 132, 2010
|
|
