8ATF
 
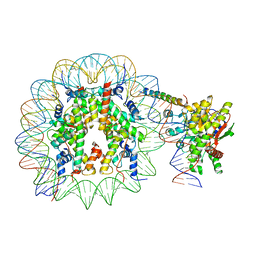 | | Nucleosome-bound Ino80 ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (226-MER), DNA (227-MER), ... | | Authors: | Kunert, F, Metzner, F.J, Eustermann, S, Jung, J, Woike, S, Schall, K, Kostrewa, D, Hopfner, K.P. | | Deposit date: | 2022-08-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural mechanism of extranucleosomal DNA readout by the INO80 complex.
Sci Adv, 8, 2022
|
|
9R04
 
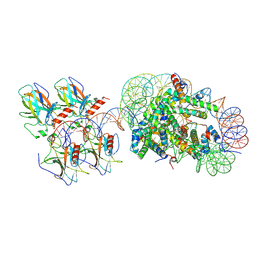 | | p53 bound to the nucleosome at position SHL-5.7 (crosslinked sample) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Michael, A.K, Kempf, G, Cavadini, S, Kater, L, Thoma, N.H. | | Deposit date: | 2025-04-24 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | p53 bound to the nucleosome at position SHL-5.7 (crosslinked sample)
To Be Published
|
|
9K3Z
 
 | |
9K40
 
 | |
9K42
 
 | |
9N6K
 
 | | 2.88 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex with DNA-binding domain | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9L1X
 
 | | hDEK-nucleosome complex (conformation 1) | | Descriptor: | 601 DNA (189-MER), 601 DNA_R (189-MER), Histone H2A type 1-B/E, ... | | Authors: | Liu, Y, Wang, C, Huang, H. | | Deposit date: | 2024-12-16 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | DEK-nucleosome structure shows DEK modulates H3K27me3 and stem cell fate.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9K41
 
 | |
9K43
 
 | |
9R2Q
 
 | | p53 bound to nucleosome at position SHL-5.7 (non-crosslinked sample) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Michael, A.K, Kempf, G, Cavadini, S, Kater, L, Thoma, N.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | p53 bound to nucleosome at position SHL-5.7 (non-crosslinked sample)
To Be Published
|
|
9R2M
 
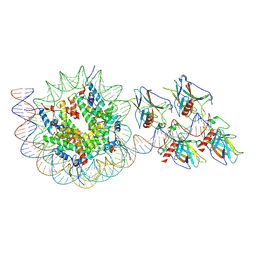 | | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | p53 bound to nucleosome at position SHL+5.9 (non-crosslinked sample, composite map)
To Be Published
|
|
9QAJ
 
 | |
9NH8
 
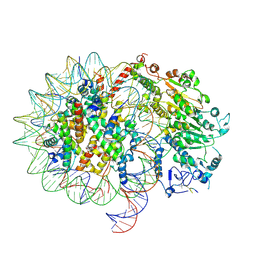 | | CHD1-nucleosome complex (anchored state) | | Descriptor: | ARGININE, Chromodomain-helicase-DNA-binding protein 1, DNA (158-MER), ... | | Authors: | James, A.M, Farnung, L. | | Deposit date: | 2025-02-24 | | Release date: | 2025-05-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human CHD1 nucleosome recruitment and pausing.
Mol.Cell, 85, 2025
|
|
9K9L
 
 | | Cryo-EM structure of the human CENP-A-H4 octasome. | | Descriptor: | Histone H3-like centromeric protein A, Histone H4, Widom601 DNA FW (145-MER), ... | | Authors: | Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2024-10-27 | | Release date: | 2025-04-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM Analysis of a Unique Subnucleosome Containing Centromere-Specific Histone Variant CENP-A.
Genes Cells, 30, 2025
|
|
9N6H
 
 | | 2.54 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 1:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9L22
 
 | | hDEK-nucleosome complex (conformation 2) | | Descriptor: | 601 DNA (189-MER), 601 DNA_R (189-MER), Histone H2A type 1-B/E, ... | | Authors: | Liu, Y, Wang, C, Huang, H. | | Deposit date: | 2024-12-16 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DEK-nucleosome structure shows DEK modulates H3K27me3 and stem cell fate.
Nat.Struct.Mol.Biol., 32, 2025
|
|
9N6I
 
 | | 2.61 A S.cerevisiae Chd1[L886G/L889G/L891G]-nucleosome 2:1 complex | | Descriptor: | Chromo domain-containing protein 1, DNA Lagging Strand, DNA Tracking Strand, ... | | Authors: | Nodelman, I.M, Folkwein, H.J, Armache, J.-P, Bowman, G.D. | | Deposit date: | 2025-02-05 | | Release date: | 2025-06-11 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | A competitive regulatory mechanism of the Chd1 remodeler is integral to distorting nucleosomal DNA.
Nat.Struct.Mol.Biol., 2025
|
|
9R2P
 
 | | p53 bound to nucleosome at position SHL+5.9 (crosslinked sample) | | Descriptor: | Cellular tumor antigen p53, DNA-for, DNA-rev, ... | | Authors: | Chakraborty, D, Kater, L, Kempf, G, Cavadini, S, Thoma, N.H. | | Deposit date: | 2025-04-30 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | p53 bound to nucleosome at position SHL+5.9 (crosslinked sample)
To Be Published
|
|
5Z23
 
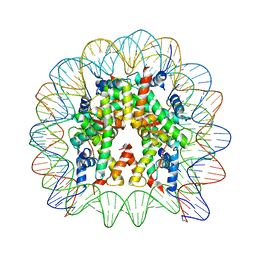 | | Crystal structure of the nucleosome containing a chimeric histone H3/CENP-A CATD | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Arimura, Y, Tachiwana, H, Takagi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2019-02-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
5Z3V
 
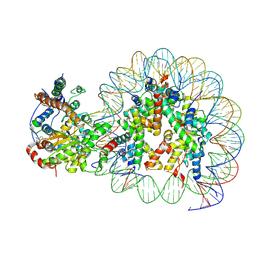 | | Structure of Snf2-nucleosome complex at shl-2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z3O
 
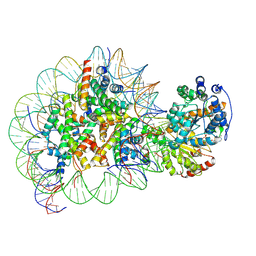 | | Structure of Snf2-nucleosome complex in ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-04-03 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
5Z30
 
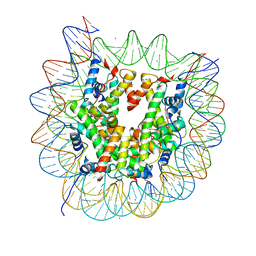 | | The crystal structure of the nucleosome containing a cancer-associated histone H2A.Z R80C mutant | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A.Z, ... | | Authors: | Horikoshi, N, Arimura, Y, Kurumizaka, H. | | Deposit date: | 2018-01-05 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Cancer-associated mutations of histones H2B, H3.1 and H2A.Z.1 affect the structure and stability of the nucleosome.
Nucleic Acids Res., 46, 2018
|
|
6A5L
 
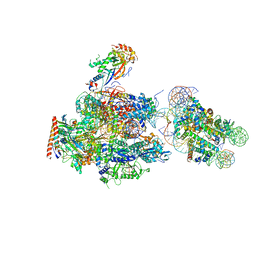 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome, with foreign DNA | | Descriptor: | DNA (198-MER), DNA (42-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5R
 
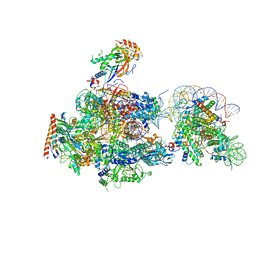 | | RNA polymerase II elongation complex stalled at SHL(-2) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
6A5T
 
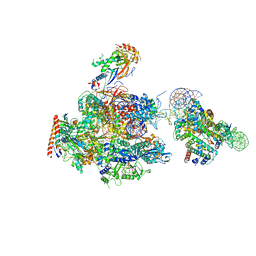 | | RNA polymerase II elongation complex stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Kujirai, T, Ehara, H, Fujino, Y, Shirouzu, M, Sekine, S, Kurumizaka, H. | | Deposit date: | 2018-06-25 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Structural basis of the nucleosome transition during RNA polymerase II passage.
Science, 362, 2018
|
|
