6HKZ
 
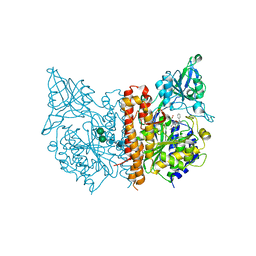 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor RNA 2-49-1 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[2-[(6-fluoranylpyridin-3-yl)carbonylamino]ethyl-phenyl-amino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
6PY1
 
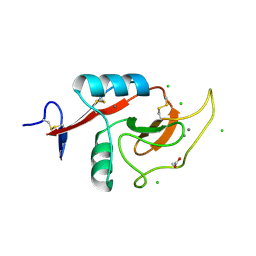 | | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, ACETATE ION, C-type lectin domain family 10 member A, ... | | Authors: | Birrane, G, Murphy, P.V, Gabba, A, Luz, J.G. | | Deposit date: | 2019-07-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Crystal Structure of the Carbohydrate Recognition Domain of the Human Macrophage Galactose C-Type Lectin Bound to GalNAc and the Tumor-Associated Tn Antigen.
Biochemistry, 60, 2021
|
|
8YEY
 
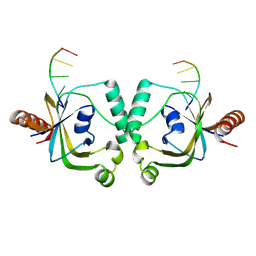 | | TRIP4 ASCH domain in complex with ssDNA-1 | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*TP*TP*TP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-02-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
8YXW
 
 | | TRIP4 ASCH domain in complex with a 12bp dsDNA (5'-TGAGGTACCTCC-3') | | Descriptor: | Activating signal cointegrator 1, DNA (5'-D(*GP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*CP*C)-3') | | Authors: | Ding, J, Yang, H, Hu, C. | | Deposit date: | 2024-04-03 | | Release date: | 2024-06-26 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of the DNA-binding properties of human TRIP4 ASCH domain reveals insights into its functional role.
Structure, 32, 2024
|
|
6RPV
 
 | | Extremely stable monomeric variant of human cystatin C with single amino acid substitution | | Descriptor: | Cystatin-C | | Authors: | Zhukov, I, Rodziewicz-Motowidlo, S, Maszota-Zieleniak, M, Jurczak, P, Kozak, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | NMR and crystallographic structural studies of the extremely stable monomeric variant of human cystatin C with single amino acid substitution.
Febs J., 287, 2020
|
|
2M75
 
 | |
8R6W
 
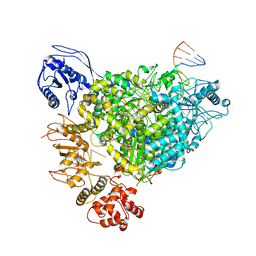 | | Structure of the SFTSV L protein in a transcription-priming state with bound capped RNA [TRANSCRIPTION-PRIMING] | | Descriptor: | RNA (5'-R(*(M7G)*AP*AP*A)-3'), RNA (5'-R(*AP*CP*AP*C)-3'), RNA (5'-R(*AP*CP*AP*CP*AP*GP*AP*GP*AP*CP*GP*CP*CP*CP*AP*G)-3'), ... | | Authors: | Williams, H.M, Thorkelsson, S.R, Vogel, D, Busch, C, Milewski, M, Cusack, S, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-11-23 | | Release date: | 2024-04-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural snapshots of phenuivirus cap-snatching and transcription.
Nucleic Acids Res., 52, 2024
|
|
4WGY
 
 | |
5LSI
 
 | | CRYSTAL STRUCTURE OF THE KINETOCHORE MIS12 COMPLEX HEAD2 SUBDOMAIN CONTAINING DSN1 AND NSL1 FRAGMENTS | | Descriptor: | Kinetochore-associated protein DSN1 homolog, Kinetochore-associated protein NSL1 homolog, SULFATE ION | | Authors: | Vetter, I.R, Petrovic, A, Keller, J, Liu, Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores.
Cell, 167, 2016
|
|
6VJP
 
 | | Structure of Staphylococcus aureus peptidoglycan O-acetyltransferase A (OatA) C-terminal catalytic domain | | Descriptor: | Acetyltransferase, SODIUM ION | | Authors: | Jones, C.J, Sychantha, D, Howell, P.L, Clarke, A.J. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.711 Å) | | Cite: | Structural basis for theO-acetyltransferase function of the extracytoplasmic domain of OatA fromStaphylococcus aureus.
J.Biol.Chem., 295, 2020
|
|
6HKJ
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a inhibitor RNA 2-19-1 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[(6-fluoranylpyridin-3-yl)amino]-1-oxidanyl-1,6-bis(oxidanylidene)hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Motlova, L, Novakova, Z, Barinka, C. | | Deposit date: | 2018-09-06 | | Release date: | 2018-12-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | 2-Aminoadipic Acid-C(O)-Glutamate Based Prostate-Specific Membrane Antigen Ligands for Potential Use as Theranostics.
ACS Med Chem Lett, 9, 2018
|
|
4YL6
 
 | |
9FRD
 
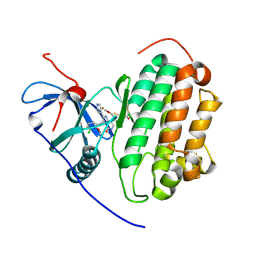 | | Wildtype EGFR bound with Compound 23 | | Descriptor: | (7~{S})-3-[(3-chloranyl-2-methoxy-phenyl)amino]-2-(3-fluoranylpyridin-4-yl)-7-(2-methoxyethyl)-1,5,6,7-tetrahydropyrrolo[3,2-c]pyridin-4-one, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CHLORIDE ION, ... | | Authors: | Pagliarini, A.R, Milgram, B.C, Borrelli, D.R, OHearn, E, Huff, M.R, Ladd, B, Brooijmans, N, Henderson, J.A, Hilbert, B.J, Wang, W, Ito, T. | | Deposit date: | 2024-06-18 | | Release date: | 2025-01-29 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of STX-721, a Covalent, Potent, and Highly Mutant-Selective EGFR/HER2 Exon20 Insertion Inhibitor for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 68, 2025
|
|
8PDP
 
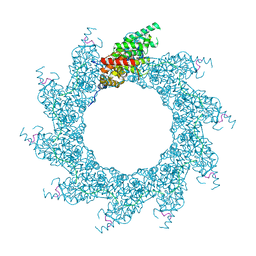 | | 10-mer ring of HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDQ
 
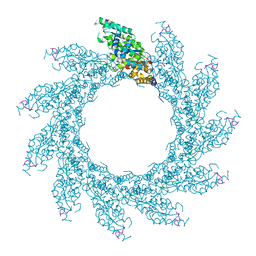 | | 11-mer ring of HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDR
 
 | | Rigid body fit of assembled HMPV N-RNA spiral bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
8PDS
 
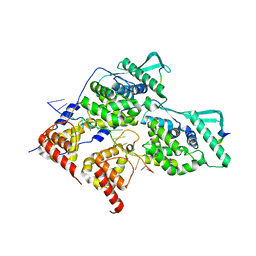 | | Local refinement of dimeric HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
1HT7
 
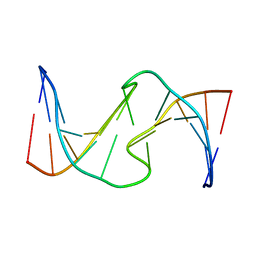 | |
6OVK
 
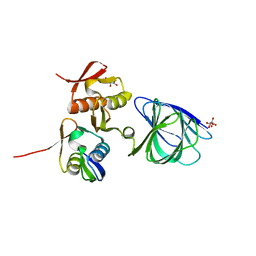 | |
4YF2
 
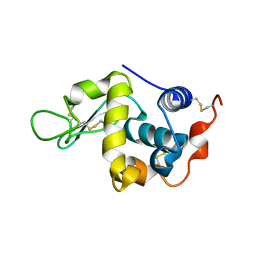 | | Crystal structure of mouse sperm C-type lysozyme-like protein 1 | | Descriptor: | Sperm acrosome membrane-associated protein 3 | | Authors: | Zheng, H, Mandal, A, Shumilin, I.A, Shabalin, I.G, Herr, J.C, Minor, W. | | Deposit date: | 2015-02-24 | | Release date: | 2015-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Sperm Lysozyme-Like Protein 1 (SLLP1), an intra-acrosomal oolemmal-binding sperm protein, reveals filamentous organization in protein crystal form.
Andrology, 3, 2015
|
|
7AU0
 
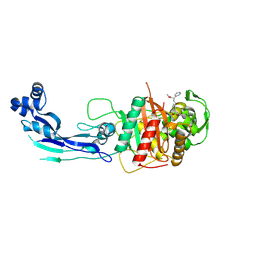 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 7) | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI, methyl (R)-2-(1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborole-6-carboxamido)-2-phenylacetate | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7ATX
 
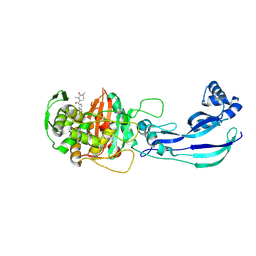 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 4) | | Descriptor: | 4-(1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborole-6-carbonyl)-1,3,3-trimethylpiperazin-2-one, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-01 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7AU8
 
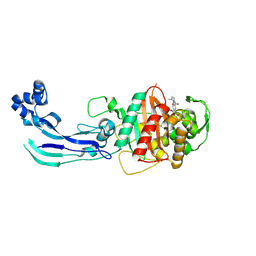 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 13) | | Descriptor: | 2-(1-hydroxy-6-((2-(4-methyl-3-oxopiperazin-1-yl)-2-oxoethyl)carbamoyl)-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acetic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7AU9
 
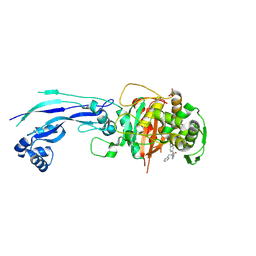 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 14) | | Descriptor: | GLYCEROL, N,N-dibenzyl-1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborole-6-carboxamide, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
7AUB
 
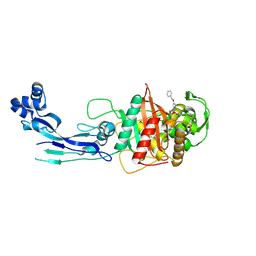 | | Structure of P. aeruginosa PBP3 in complex with a benzoxaborole (Compound 15) | | Descriptor: | 2-(5-(benzyloxy)-1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acetic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Newman, H, Bellini, B, Dowson, C.G. | | Deposit date: | 2020-11-02 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.907 Å) | | Cite: | High-Throughput Crystallography Reveals Boron-Containing Inhibitors of a Penicillin-Binding Protein with Di- and Tricovalent Binding Modes.
J.Med.Chem., 64, 2021
|
|
