8YIH
 
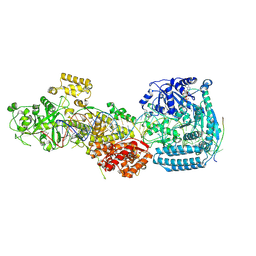 | | DmDcr-2/LoqsPD/slm1 in pre-dicing state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-2, isoform A, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
8YIG
 
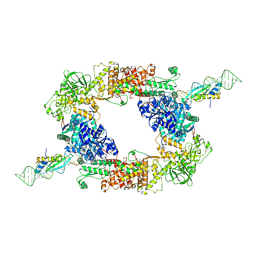 | | DmDcr-2/LoqsPD/slm2 in initial binding state | | Descriptor: | Dicer-2, isoform A, Isoform PD of Protein Loquacious, ... | | Authors: | Cao, N, Su, S, Wang, J, Ma, J, Wang, H.-W. | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of endo-siRNA processing by Drosophila Dicer-2 and Loqs-PD.
Nucleic Acids Res., 53, 2025
|
|
7SN6
 
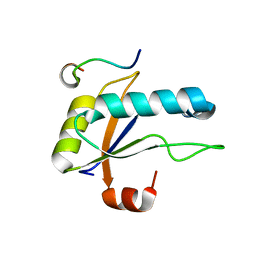 | | U2AF65 UHM BOUND TO SF3B155 ULM5 | | Descriptor: | ISOPROPYL ALCOHOL, Splicing factor 3B subunit 1, Splicing factor U2AF 65 kDa subunit | | Authors: | Loerch, S, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2021-10-27 | | Release date: | 2022-07-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A UHM-ULM interface with unusual structural features contributes to U2AF2 and SF3B1 association for pre-mRNA splicing.
J.Biol.Chem., 298, 2022
|
|
7KFN
 
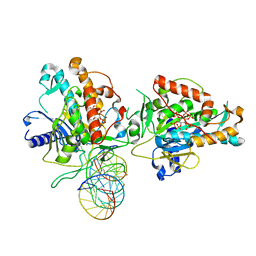 | | Structure of Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA containing a 2'-deoxy Benner's Base Z opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, Gli1 1W5 23mer RNA, Gli1 8AZ 23mer RNA, ... | | Authors: | Wilcox, X.E, Fisher, A.J, Beal, P.A. | | Deposit date: | 2020-10-14 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rational Design of RNA Editing Guide Strands: Cytidine Analogs at the Orphan Position.
J.Am.Chem.Soc., 143, 2021
|
|
1FYO
 
 | |
3AEV
 
 | | Crystal structure of a/eIF2alpha-aDim2p-rRNA complex from Pyrococcus horikoshii OT3 | | Descriptor: | Putative uncharacterized protein PH1566, RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*CP*UP*CP*C)-3'), Translation initiation factor 2 subunit alpha | | Authors: | Tanokura, M, Jia, M.Z, Nagata, K. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An archaeal Dim2-like protein, aDim2p, forms a ternary complex with a/eIF2 alpha and the 3' end fragment of 16S rRNA
J.Mol.Biol., 398, 2010
|
|
7U4A
 
 | | Crystal Structure of Zika virus xrRNA1 mutant | | Descriptor: | MAGNESIUM ION, RNA (70-MER) | | Authors: | Thompson, R.D, Carbaugh, D.L, Nielsen, J.R, Witt, C, Meganck, R.M, Rangadurai, A, Zhao, B, Bonin, J.P, Nathan, N.T, Marzluff, W.F, Frank, A.T, Lazear, H.M, Zhang, Q. | | Deposit date: | 2022-02-28 | | Release date: | 2023-09-06 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Lifetime of ground conformational state determines the activity of structured RNA.
Nat.Chem.Biol., 2025
|
|
3OVA
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-adding enzyme, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OVS
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OVB
 
 | | How the CCA-adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-16 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OUY
 
 | | How the CCA-adding Enzyme Selects Adenine Over Cytosine at Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, CCA-Adding Enzyme, PYROPHOSPHATE 2-, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
3OV7
 
 | | How the CCA-Adding Enzyme Selects Adenine over Cytosine in Position 76 of tRNA | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CCA-Adding Enzyme, ... | | Authors: | Pan, B.C, Xiong, Y, Steitz, T.A. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How the CCA-Adding Enzyme Selects Adenine over Cytosine at Position 76 of tRNA.
Science, 330, 2010
|
|
4MDX
 
 | |
1OB5
 
 | | T. aquaticus elongation factor EF-Tu complexed with the antibiotic enacyloxin IIa, a GTP analog, and Phe-tRNA | | Descriptor: | ELONGATION FACTOR TU, ENACYLOXIN IIA, MAGNESIUM ION, ... | | Authors: | Dahlberg, C, Nielsen, R.C, Parmeggiani, A, Nyborg, J, Nissen, P. | | Deposit date: | 2003-01-24 | | Release date: | 2005-10-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Enacyloxin Iia Pinpoints a Binding Pocket of Elongation Factor TU for Development of Novel Antibiotics.
J.Biol.Chem., 281, 2006
|
|
1FT8
 
 | |
4GHA
 
 | | Crystal structure of Marburg virus VP35 RNA binding domain bound to 12-bp dsRNA | | Descriptor: | Polymerase cofactor VP35, RNA (5'-R(*CP*UP*AP*GP*AP*CP*GP*UP*CP*UP*AP*G)-3') | | Authors: | Bale, S, Jean-Philippe, J, Bornholdt, Z.A, Kimberlin, C.K, Halfmann, P, Zandonatti, M.A, Kunert, J, Kroon, G.J.A, Kawaoka, Y, MacRae, I.J, Wilson, I.A, Saphire, E.O. | | Deposit date: | 2012-08-07 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Marburg Virus VP35 Can Both Fully Coat the Backbone and Cap the Ends of dsRNA for Interferon Antagonism.
Plos Pathog., 8, 2012
|
|
4M59
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 in complex with an 18-nt psaJ RNA element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Yin, P, Li, Q, Yan, C, Liu, Y, Yan, N. | | Deposit date: | 2013-08-08 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the modular recognition of single-stranded RNA by PPR proteins.
Nature, 504, 2013
|
|
8XX4
 
 | | ASFV RNAP elongation complex | | Descriptor: | C122R, D339L, DNA (5'-D(P*CP*GP*CP*AP*AP*CP*GP*GP*CP*GP*A)-3'), ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8S8W
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8S8X
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
7LD5
 
 | | polynucleotide phosphorylase | | Descriptor: | MAGNESIUM ION, Polyribonucleotide nucleotidyltransferase, poly-A RNA fragment | | Authors: | Goldgur, Y, Shuman, S, De La Cruz, M.J, Ghosh, S, Unciuleac, M.-C. | | Deposit date: | 2021-01-12 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure and mechanism of Mycobacterium smegmatis polynucleotide phosphorylase.
Rna, 27, 2021
|
|
9GS9
 
 | | Tn7016 PseCAST QCascade | | Descriptor: | Cas6, Cas7.1, Cas8, ... | | Authors: | Lampe, G.D, Liang, A.R, Zhang, D.J, Fernandez, I.S, Sternberg, S.H. | | Deposit date: | 2024-09-13 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-guided engineering of type I-F CASTs for targeted gene insertion in human cells.
Biorxiv, 2024
|
|
9CGV
 
 | | SARS-CoV-2 nsp12 NiRAN domain bound to a covalent inhibitor SW090466-1 | | Descriptor: | MANGANESE (II) ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Osinski, A, Hernandez, G, Tagliabracci, V.S. | | Deposit date: | 2024-07-01 | | Release date: | 2025-03-12 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Covalent inhibition of the SARS-CoV-2 NiRAN domain via an active-site cysteine.
J.Biol.Chem., 301, 2025
|
|
9E9Q
 
 | |
7KKV
 
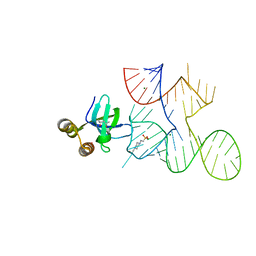 | | Crystal structure of Bacillus halodurans OapB in complex with its OLE RNA target (native, crystal form I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, MAGNESIUM ION, ... | | Authors: | Yang, Y, Breaker, R.R. | | Deposit date: | 2020-10-28 | | Release date: | 2021-03-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a bacterial OapB protein with its OLE RNA target gives insights into the architecture of the OLE ribonucleoprotein complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
