5QLI
 
 | |
5QLW
 
 | |
5QMC
 
 | |
5QMU
 
 | |
5QN5
 
 | |
5H30
 
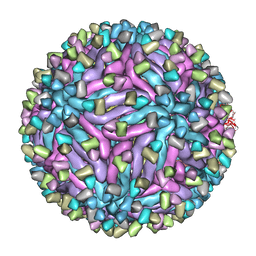 | | Cryo-EM structure of zika virus complexed with Fab C10 at pH 6.5 | | Descriptor: | IgG C10 heavy chain, IgG C10 light chain, structural protein E, ... | | Authors: | Zhang, S, Kostyuchenko, V, Ng, T.-S, Lok, S.-M. | | Deposit date: | 2016-10-19 | | Release date: | 2016-11-30 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Neutralization mechanism of a highly potent antibody against Zika virus
Nat Commun, 7, 2016
|
|
4DYN
 
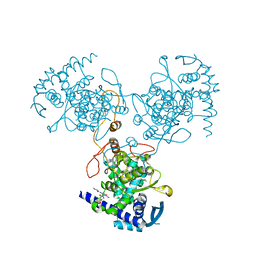 | | Crystal Structure of WSN/A Influenza Nucleoprotein with BMS-885838 Ligand Bound | | Descriptor: | N-[4-chloranyl-5-[4-[[3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]carbonyl]piperazin-1-yl]-2-nitro-phenyl]pyridine-2-carboxamide, Nucleocapsid protein | | Authors: | Lewis, H.A, Baldwin, E.T, Steinbacher, S, Maskos, K, Mortl, M, Kiefersauer, R, Edavettal, S, McDonnell, P.A, Pearce, B.C, Langley, D.R. | | Deposit date: | 2012-02-29 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | To be determined
To be Published
|
|
5QKG
 
 | |
5QKW
 
 | |
5QLF
 
 | |
5QLP
 
 | |
5QM4
 
 | |
5QMK
 
 | |
5QMZ
 
 | |
5QNE
 
 | |
5QKE
 
 | |
5QKV
 
 | |
5QLB
 
 | |
3LXG
 
 | | Crystal structure of rat phosphodiesterase 10A in complex with ligand WEB-3 | | Descriptor: | 2-methoxy-6,7-dimethyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Mosbacher, T, Jestel, A, Steinbacher, S. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of imidazo[1,5-a]pyrido[3,2-e]pyrazines as a new class of phosphodiesterase 10A inhibitiors.
J.Med.Chem., 53, 2010
|
|
4YLQ
 
 | | Crystal Structure of a FVIIa-Trypsin Chimera (FT) in Complex with Soluble Tissue Factor | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-PROPANOL, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity.
J.Biol.Chem., 291, 2016
|
|
6UTD
 
 | |
6B2W
 
 | | C. Jejuni C315S Agmatine Deiminase with Substrate Bound | | Descriptor: | AGMATINE, POTASSIUM ION, Putative peptidyl-arginine deiminase family protein | | Authors: | Shek, R, Hicks, K.A, French, J.B. | | Deposit date: | 2017-09-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Basis for Targeting Campylobacter jejuni Agmatine Deiminase To Overcome Antibiotic Resistance.
Biochemistry, 56, 2017
|
|
6ANK
 
 | | Synaptotagmin-7, C2A- and C2B-domains | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4RJT
 
 | |
5OIN
 
 | | InhA (T2A mutant) complexed with N-(1-(pyrimidin-2-yl)piperidin-4-yl)acetamide | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ~{N}-(1-pyrimidin-2-ylpiperidin-4-yl)ethanamide | | Authors: | Convery, M.A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Screening of a Novel Fragment Library with Functional Complexity against Mycobacterium tuberculosis InhA.
ChemMedChem, 13, 2018
|
|
