7T78
 
 | |
6U00
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
8GRJ
 
 | | Crystal structure of gamma-alpha subunit complex from Burkholderia cepacia FAD glucose dehydrogenase in complex with gluconolactone | | Descriptor: | D-glucono-1,5-lactone, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Yoshida, H, Kojima, K, Tsugawa, W, Okuda-Shimazaki, J, Kerrigan, J.A, Sode, K. | | Deposit date: | 2022-09-01 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Improvement of substrate specificity of the direct electron transfer type FAD-dependent glucose dehydrogenase catalytic subunit.
J.Biotechnol., 2024
|
|
6U05
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-13 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
7R70
 
 | | Crystal Structure of the UbArk2C fusion protein | | Descriptor: | GLYCEROL, Ubiquitin,E3 ubiquitin-protein ligase RNF165, ZINC ION | | Authors: | Paluda, A, Middleton, A.J, Mace, P.D, Day, C.L. | | Deposit date: | 2021-06-24 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ubiquitin and a charged loop regulate the ubiquitin E3 ligase activity of Ark2C.
Nat Commun, 13, 2022
|
|
9M3N
 
 | | Crystal structure of human TRIM21 PRYSPRY in complex with T-02 | | Descriptor: | 1-ethyl-~{N}-methyl-5-phenyl-~{N}-[3-[3-(trifluoromethyl)phenyl]cyclobutyl]pyrazole-4-carboxamide, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Zhang, L.J, Zhang, L.Y. | | Deposit date: | 2025-03-03 | | Release date: | 2025-07-16 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | TRIM21-Driven Degradation of BRD4: Development of Heterobifunctional Degraders and Investigation of Recruitment and Selectivity Mechanisms.
J.Chem.Inf.Model., 2025
|
|
6TZO
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
7QRX
 
 | |
3PZC
 
 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with Coenzyme A | | Descriptor: | ACETATE ION, Amino acid--[acyl-carrier-protein] ligase 1, COENZYME A, ... | | Authors: | Weygand-Durasevic, I, Luic, M, Mocibob, M, Ivic, N, Subasic, D. | | Deposit date: | 2010-12-14 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substrate Recognition by Novel Family of Amino Acid:[Carrier Protein] Ligases
Croatica Chemica Acta, 84, 2011
|
|
2VUG
 
 | | The structure of an archaeal homodimeric RNA ligase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PAB1020, ... | | Authors: | Brooks, M.A, Meslet-Cladiere, L, Graille, M, Kuhn, J, Blondeau, K, Myllykallio, H, van Tilbeurgh, H. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of an archaeal homodimeric ligase which has RNA circularization activity.
Protein Sci., 17, 2008
|
|
6TZM
 
 | | Crystal Structure of Fungal RNA Kinase | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, tRNA ligase | | Authors: | Shuman, S, Goldgur, Y, Banerjee, A. | | Deposit date: | 2019-08-12 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Atomic structures of the RNA end-healing 5'-OH kinase and 2',3'-cyclic phosphodiesterase domains of fungal tRNA ligase: conformational switches in the kinase upon binding of the GTP phosphate donor.
Nucleic Acids Res., 47, 2019
|
|
3PT3
 
 | | Crystal structure of the C-terminal lobe of the human UBR5 HECT domain | | Descriptor: | E3 ubiquitin-protein ligase UBR5 | | Authors: | Matta-Camacho, E, Kozlov, G, Menade, M, Gehring, K. | | Deposit date: | 2010-12-02 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the HECT C-lobe of the UBR5 E3 ubiquitin ligase.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4EGP
 
 | | The X-ray crystal structure of CYP199A4 in complex with 2-naphthoic acid | | Descriptor: | CHLORIDE ION, Cytochrome P450, GLYCEROL, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
4TRH
 
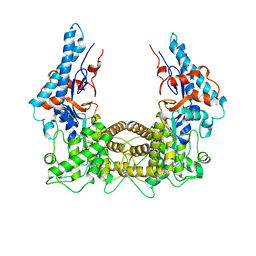 | | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling | | Descriptor: | SidC | | Authors: | Hsu, F.S, Luo, X, Qiu, J, Teng, Y, Jin, J, Smolka, M.B, Luo, Z.Q, Mao, Y. | | Deposit date: | 2014-06-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The Legionella effector SidC defines a unique family of ubiquitin ligases important for bacterial phagosomal remodeling.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2A0Z
 
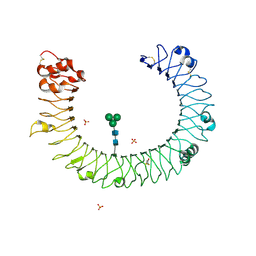 | | The molecular structure of toll-like receptor 3 ligand binding domain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bell, J.K, Botos, I, Hall, P.R, Askins, J, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2005-06-17 | | Release date: | 2005-08-02 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The molecular structure of the Toll-like receptor 3 ligand-binding domain
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3NIH
 
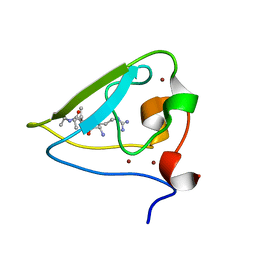 | | The structure of UBR box (RIAAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide RIAAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3E5N
 
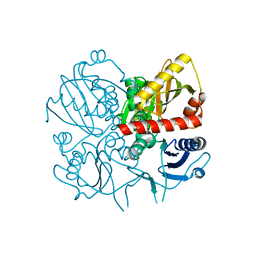 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
4A0K
 
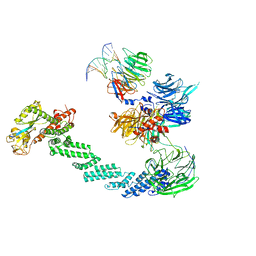 | | STRUCTURE OF DDB1-DDB2-CUL4A-RBX1 BOUND TO A 12 BP ABASIC SITE CONTAINING DNA-DUPLEX | | Descriptor: | 12 BP DNA, 12 BP THF CONTAINING DNA, CULLIN-4A, ... | | Authors: | Fischer, E.S, Scrima, A, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-09 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (5.93 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation.
Cell(Cambridge,Mass.), 147, 2011
|
|
3FL2
 
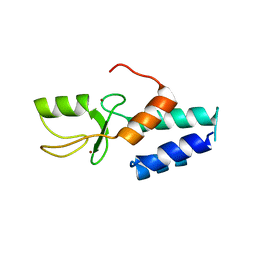 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
3NIL
 
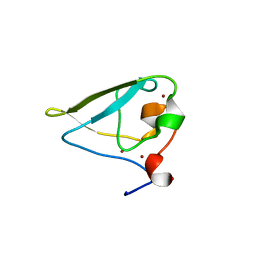 | | The structure of UBR box (RDAA) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, Peptide RDAA, ... | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIK
 
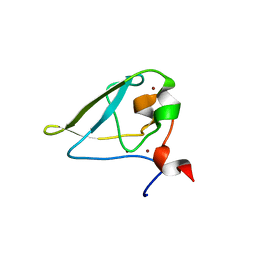 | | The structure of UBR box (REAA) | | Descriptor: | E3 ubiquitin-protein ligase UBR1, Peptide REAA, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NIS
 
 | | The structure of UBR box (native2) | | Descriptor: | ACETATE ION, E3 ubiquitin-protein ligase UBR1, ZINC ION | | Authors: | Choi, W.S, Jeong, B.-C, Lee, M.-R, Song, H.K. | | Deposit date: | 2010-06-16 | | Release date: | 2010-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases
Nat.Struct.Mol.Biol., 17, 2010
|
|
2PZD
 
 | |
4UCU
 
 | | Fragment bound to H.influenza NAD dependent DNA ligase | | Descriptor: | 1-(2,4-dimethylbenzyl)-6-oxo-1,6-dihydropyridine-3-carboxamide, 8-hydroxyquinoline-2-carboxylic acid, DNA LIGASE | | Authors: | Hale, M, Brassington, C, Carcanague, D, Embrey, K, Eyermann, C.J, Giacobbe, R.A, Gingipali, L, Gowravaram, M, Harang, J, Howard, T, Ioannidis, G, Jahic, H, Kutschke, A, Laganas, V.A, Loch, J, Miller, M.D, Murphy-Benenato, K.E, Oguto, H, Otterbein, L, Patel, S.J, Shapiro, A.B, Boriack-Sjodin, P.A. | | Deposit date: | 2014-12-04 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | From Fragments to Leads: Novel Bacterial Nad+-Dependent DNA Ligase Inhibitors
Tetrahedron Lett., 56, 2015
|
|
2YZ1
 
 | |
