2A53
 
 | | fluorescent protein asFP595, A143S, off-state | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A54
 
 | | fluorescent protein asFP595, A143S, on-state, 1min irradiation | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A55
 
 | | Solution structure of the two N-terminal CCP modules of C4b-binding protein (C4BP) alpha-chain. | | Descriptor: | C4b-binding protein | | Authors: | Jenkins, H.T, Mark, L, Ball, G, Lindahl, G, Uhrin, D, Blom, A.M, Barlow, P.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Human C4b-binding Protein, Structural Basis for Interaction with Streptococcal M Protein, a Major Bacterial Virulence Factor
J.Biol.Chem., 281, 2006
|
|
2A56
 
 | | fluorescent protein asFP595, A143S, on-state, 5min irradiation | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A57
 
 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A58
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound riboflavin | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A59
 
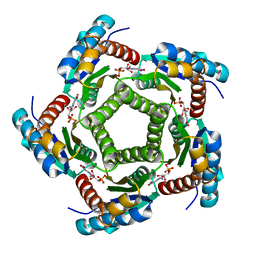 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A5A
 
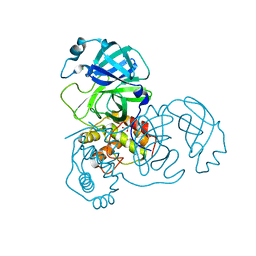 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5B
 
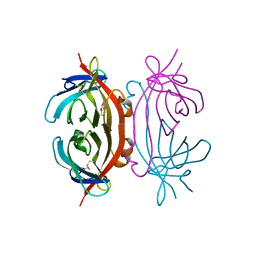 | | Avidin complexed with 8-oxodeoxyguanosine | | Descriptor: | 2'-DEOXY-8-OXOGUANOSINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
2A5C
 
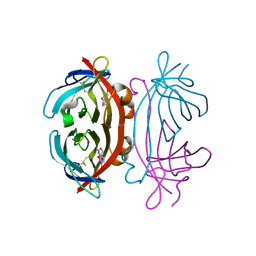 | | Structure of Avidin in complex with the ligand 8-oxodeoxyadenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-OXODEOXYADENOSINE, Avidin | | Authors: | Conners, R, Hooley, E, Thomas, S, Brady, R.L. | | Deposit date: | 2005-06-30 | | Release date: | 2006-05-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recognition of oxidatively modified bases within the biotin-binding site of avidin.
J.Mol.Biol., 357, 2006
|
|
2A5D
 
 | | Structural basis for the activation of cholera toxin by human ARF6-GTP | | Descriptor: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | Authors: | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
2A5E
 
 | | SOLUTION NMR STRUCTURE OF TUMOR SUPPRESSOR P16INK4A, RESTRAINED MINIMIZED MEAN STRUCTURE | | Descriptor: | TUMOR SUPPRESSOR P16INK4A | | Authors: | Byeon, I.-J.L, Li, J, Ericson, K, Selby, T.L, Tevelev, A, Kim, H.-J, O'Maille, P, Tsai, M.-D. | | Deposit date: | 1998-02-13 | | Release date: | 1999-08-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Tumor suppressor p16INK4A: determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4.
Mol.Cell, 1, 1998
|
|
2A5F
 
 | | Cholera toxin A1 subunit bound to its substrate, NAD+, and its human protein activator, ARF6 | | Descriptor: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | Authors: | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
2A5G
 
 | | Cholera toxin A1 subunit bound to ARF6(Q67L) | | Descriptor: | ADP-ribosylation factor 6, Cholera enterotoxin, A chain, ... | | Authors: | O'Neal, C.J, Jobling, M.G, Holmes, R.K, Hol, W.G.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis for the activation of cholera toxin by human ARF6-GTP.
Science, 309, 2005
|
|
2A5H
 
 | | 2.1 Angstrom X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale SB4, with Michaelis analog (L-alpha-lysine external aldimine form of pyridoxal-5'-phosphate). | | Descriptor: | IRON/SULFUR CLUSTER, L-lysine 2,3-aminomutase, LYSINE, ... | | Authors: | Lepore, B.W, Ruzicka, F.J, Frey, P.A, Ringe, D. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-04 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The X-ray crystal structure of lysine-2,3-aminomutase from Clostridium subterminale.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A5I
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5J
 
 | | Crystal Structure of Human RAB2B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-2B | | Authors: | Dong, A, Wang, J, Shen, Y, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Bochkarev, A, Park, H.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Crystal structure of human RAB2B
To be Published
|
|
2A5K
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2A5L
 
 | | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa | | Descriptor: | MAGNESIUM ION, Trp repressor binding protein WrbA | | Authors: | Lunin, V.V, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of the Trp repressor binding protein WrbA from Pseudomonas aeruginosa
To be Published
|
|
2A5M
 
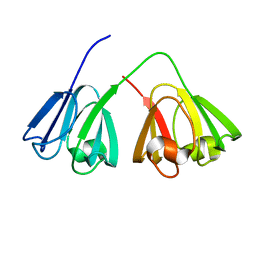 | |
2A5P
 
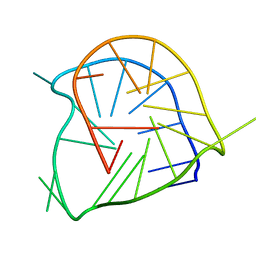 | | Monomeric parallel-stranded DNA tetraplex with snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, NMR, 8 struct. | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
2A5R
 
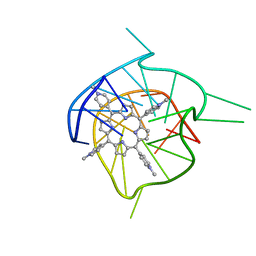 | | Complex of tetra-(4-n-methylpyridyl) porphin with monomeric parallel-stranded DNA tetraplex, snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, C-MYC promoter, NMR, 6 struct. | | Descriptor: | (1Z,4Z,9Z,15Z)-5,10,15,20-tetrakis(1-methylpyridin-1-ium-4-yl)-21,23-dihydroporphyrin, 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
2A5S
 
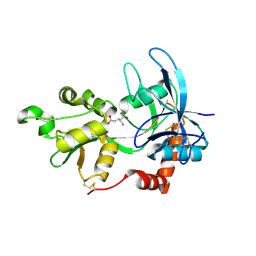 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5T
 
 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5U
 
 | | Crystal Structure of human PI3Kgamma complexed with AS605240 | | Descriptor: | (5E)-5-(QUINOXALIN-6-YLMETHYLENE)-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-07-01 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
