4A2S
 
 | | Structure of the engineered retro-aldolase RA95.5 | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, INDOLE-3-GLYCEROL PHOSPHATE SYNTHASE | | Authors: | Giger, L, Caner, S, Kast, P, Baker, D, Ban, N, Hilvert, D. | | Deposit date: | 2011-09-28 | | Release date: | 2012-11-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Evolution of a designed retro-aldolase leads to complete active site remodeling.
Nat.Chem.Biol., 9, 2013
|
|
2V0L
 
 | | Characterization of Substrate Binding and Catalysis of the Potential Antibacterial Target N-acetylglucosamine-1-phosphate Uridyltransferase (GlmU) | | Descriptor: | BIFUNCTIONAL PROTEIN GLMU, SULFATE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Mochalkin, I, Lightle, S, Ohren, J.F, Chirgadze, N.Y. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Substrate Binding and Catalysis in the Potential Antibacterial Target N-Acetylglucosamine-1-Phosphate Uridyltransferase (Glmu).
Protein Sci., 16, 2007
|
|
7HCX
 
 | |
7HFY
 
 | |
3RIH
 
 | |
2UV2
 
 | | Crystal Structure Of Human Ste20-Like Kinase Bound To 4-(4-(5- Cyclopropyl-1H-pyrazol-3-ylamino)-quinazolin-2-ylamino)-phenyl)- acetonitrile | | Descriptor: | 1,2-ETHANEDIOL, STE20-LIKE SERINE-THREONINE KINASE, THIOCYANATE ION, ... | | Authors: | Pike, A.C.W, Rellos, P, Fedorov, O, Keates, T, Salah, E, Savitsky, P, Papagrigoriou, E, Bunkoczi, G, Debreczeni, J.E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-03-08 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Activation Segment Dimerization: A Mechanism for Kinase Autophosphorylation of Non-Consensus Sites.
Embo J., 27, 2008
|
|
1VKE
 
 | |
7HF1
 
 | |
3R21
 
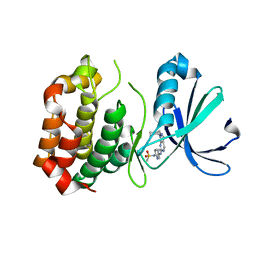 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3FJA
 
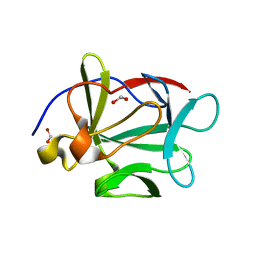 | |
4FBE
 
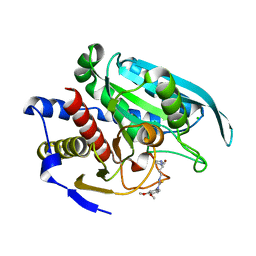 | | Crystal structure of the C136A/C164A variant of mitochondrial isoform of glutaminyl cyclase from Drosophila melanogaster | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, CG5976, isoform B, ... | | Authors: | Kolenko, P, Koch, B, Ruiz-Carilo, D, Stubbs, M.T. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of Glutaminyl Cyclases (QCs) from Drosophila melanogaster Reveal Active Site Conservation between Insect and Mammalian QCs.
Biochemistry, 51, 2012
|
|
4OUI
 
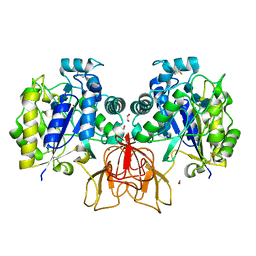 | | Structure of Vibrio cholerae chitin de-N-acetylase in complex with TRIACETYLCHITOTRIOSE (CTO) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Deacetylase DA1, ... | | Authors: | Albesa-Jove, D, Andres, E, Biarnes, X, Planas, A, Guerin, M.E. | | Deposit date: | 2014-02-17 | | Release date: | 2014-08-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structural basis of chitin oligosaccharide deacetylation.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1R4Z
 
 | | Bacillus subtilis lipase A with covalently bound Rc-IPG-phosphonate-inhibitor | | Descriptor: | Lipase, [(4R)-2,2-DIMETHYL-1,3-DIOXOLAN-4-YL]METHYL HYDROGEN HEX-5-ENYLPHOSPHONATE | | Authors: | Droege, M.J, Van Pouderoyen, G, Vrenken, T.E, Rueggeberg, C.J, Reetz, M.T, Dijkstra, B.W, Quax, W.J. | | Deposit date: | 2003-10-09 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Directed Evolution of Bacillus subtilis Lipase A by Use of Enantiomeric Phosphonate Inhibitors: Crystal Structures and Phage Display Selection
Chembiochem, 7, 2005
|
|
5QR5
 
 | | PanDDA analysis group deposition -- Crystal Structure of human ALAS2A in complex with Z2241115980 | | Descriptor: | 1-[(furan-2-yl)methyl]-4-(methylsulfonyl)piperazine, 5-aminolevulinate synthase, erythroid-specific, ... | | Authors: | Bezerra, G.A, Foster, W, Bailey, H, Shrestha, L, Krojer, T, Talon, R, Brandao-Neto, J, Douangamath, A, Nicola, B.B, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Brennan, P.E, Yue, W.W. | | Deposit date: | 2019-05-22 | | Release date: | 2019-08-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5QOU
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z296300542 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4KHX
 
 | |
3WS9
 
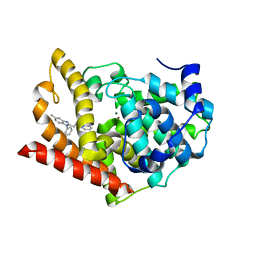 | | Crystal structure of PDE10A in complex with a benzimdazole inhibitor | | Descriptor: | 7-[2-(5-methyl-1-phenyl-1H-benzimidazol-2-yl)ethyl]imidazo[1,5-b]pyridazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-04 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Novel benzimidazole derivatives as phosphodiesterase 10A (PDE10A) inhibitors with improved metabolic stability.
Bioorg.Med.Chem., 22, 2014
|
|
7HTT
 
 | | PanDDA analysis group deposition -- Crystal Structure of FatA in complex with Z56960248 | | Descriptor: | 1-[(2R)-2,3-dihydro-1,4-benzodioxin-2-yl]-N-methylmethanamine, Oleoyl-acyl carrier protein thioesterase 1, chloroplastic | | Authors: | Kot, E, Ni, X, Tomlinson, C.W.E, Fearon, D, Aschenbrenner, J.C, Fairhead, M, Koekemoer, L, Marx, M.L, Wright, N.D, Mulholland, N.P, Montgomery, M.G, von Delft, F. | | Deposit date: | 2024-12-23 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4FJ6
 
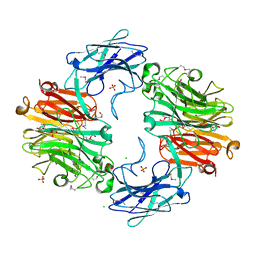 | |
2X6D
 
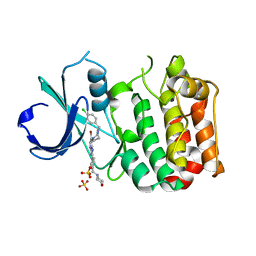 | | Aurora-A bound to an inhibitor | | Descriptor: | 6-BROMO-7-[4-(4-CHLOROBENZYL)PIPERAZIN-1-YL]-2-[4-(MORPHOLIN-4-YLMETHYL)PHENYL]-3H-IMIDAZO[4,5-B]PYRIDINE, SERINE/THREONINE-PROTEIN KINASE 6, SULFATE ION | | Authors: | Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2010-02-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Imidazo[4,5-b]pyridine derivatives as inhibitors of Aurora kinases: lead optimization studies toward the identification of an orally bioavailable preclinical development candidate.
J. Med. Chem., 53, 2010
|
|
3FJ8
 
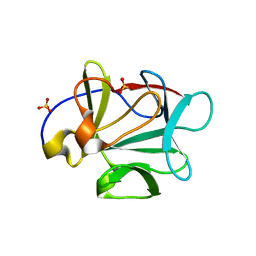 | |
2XB6
 
 | | Revisited crystal structure of Neurexin1beta-Neuroligin4 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Leone, P, Comoletti, D, Ferracci, G, Conrod, S, Garcia, S.U, Taylor, P, Bourne, Y, Marchot, P. | | Deposit date: | 2010-04-07 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into the Exquisite Selectivity of Neurexin-Neuroligin Synaptic Interactions
Embo J., 29, 2010
|
|
5YR0
 
 | | Structure of Beclin1-UVRAG coiled coil domain complex | | Descriptor: | Beclin-1, UV radiation resistance associated protein | | Authors: | Pan, X, Zhao, Y, He, Y. | | Deposit date: | 2017-11-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5QOR
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with Z1203490773 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DCP2 (NUDT20), ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
3NMR
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-22 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
