3R63
 
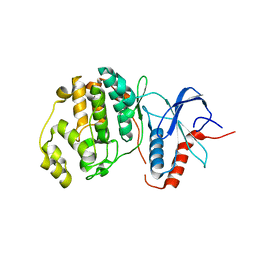 | | Structure of ERK2 (SPE) mutant (S246E) | | Descriptor: | Mitogen-activated protein kinase 1 | | Authors: | Livnah, O, Karamansha, Y. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Nuclear ERK translocation is mediated by protein kinase CK2 and accelerated by autophosphorylation.
Mol.Cell.Biol., 31, 2011
|
|
2PP4
 
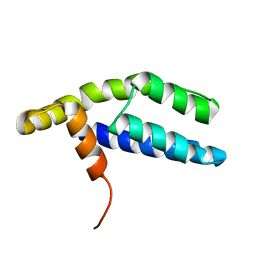 | | Solution Structure of ETO-TAFH refined in explicit solvent | | Descriptor: | Protein ETO | | Authors: | Wei, Y, Liu, S, Lausen, J, Woodrell, C, Cho, S, Biris, N, Kobayashi, N, Yokoyama, S, Werner, M.H. | | Deposit date: | 2007-04-27 | | Release date: | 2007-06-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A TAF4-homology domain from the corepressor ETO is a docking platform for positive and negative regulators of transcription
Nat.Struct.Mol.Biol., 14, 2007
|
|
2CRU
 
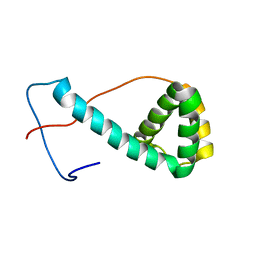 | | Solution structure of programmed cell death 5 | | Descriptor: | Programmed cell death protein 5 | | Authors: | Nagashima, T, Izumi, K, Hayashi, F, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-20 | | Release date: | 2005-11-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of programmed cell death 5
To be published
|
|
1FAV
 
 | | THE STRUCTURE OF AN HIV-1 SPECIFIC CELL ENTRY INHIBITOR IN COMPLEX WITH THE HIV-1 GP41 TRIMERIC CORE | | Descriptor: | HIV-1 ENVELOPE PROTEIN CHIMERA, PROTEIN (TRANSMEMBRANE GLYCOPROTEIN) | | Authors: | Zhou, G, Ferrer, M, Chopra, R, Strassmaier, T, Weissenhorn, W, Skehel, J.J, Oprian, D, Schreiber, S.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2000-07-13 | | Release date: | 2000-08-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of an HIV-1 specific cell entry inhibitor in complex with the HIV-1 gp41 trimeric core.
Bioorg.Med.Chem., 8, 2000
|
|
2VFJ
 
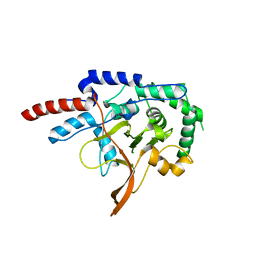 | |
2E4L
 
 | | Thermodynamic and Structural Analysis of Thermolabile RNase HI from Shewanella oneidensis MR-1 | | Descriptor: | Ribonuclease HI | | Authors: | Tadokoro, T, You, D.J, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-12-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, thermodynamic, and mutational analyses of a psychrotrophic RNase HI.
Biochemistry, 46, 2007
|
|
2DFE
 
 | | Crystal structure of Tk-RNase HII(1-200)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
2DFF
 
 | | Crystal structure of Tk-RNase HII(1-204)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
8CA0
 
 | |
1U9H
 
 | |
5AWE
 
 | | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains | | Descriptor: | Putative acetoin utilization protein, acetoin dehydrogenase | | Authors: | Nakabayashi, M, Shibata, N, Kanagawa, M, Nakagawa, N, Kuramitsu, S, Higuchi, Y. | | Deposit date: | 2015-07-03 | | Release date: | 2016-05-18 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a hypothetical protein, TTHA0829 from Thermus thermophilus HB8, composed of cystathionine-beta-synthase (CBS) and aspartate-kinase chorismate-mutase tyrA (ACT) domains.
Extremophiles, 20, 2016
|
|
6O75
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with pppApA | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, MANGANESE (II) ION, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
1U9G
 
 | | Heterocyclic Peptide Backbone Modification in GCN4-pLI Based Coiled Coils: Replacement of K(8)L(9) | | Descriptor: | General control protein GCN4, SULFATE ION | | Authors: | Horne, W.S, Yadav, M.K, Stout, C.D, Ghadiri, M.R. | | Deposit date: | 2004-08-09 | | Release date: | 2004-11-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Heterocyclic peptide backbone modifications in an alpha-helical coiled coil.
J.Am.Chem.Soc., 126, 2004
|
|
6O7B
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with cA4 | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, Cyclic RNA cA4 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
5BNO
 
 | | Crystal structure of human enterovirus D68 in complex with 6'SLN | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
5AVM
 
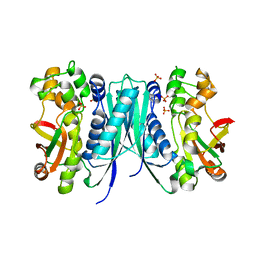 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
5FB5
 
 | |
7WP3
 
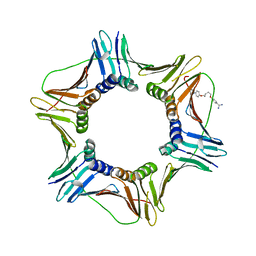 | | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, Proliferating cell nuclear antigen | | Authors: | Ahmad, M.I, Yadav, S.P, Singh, P.K, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2022-01-22 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Crystal structure of the complex of proliferating cell nuclear antigen (PCNA) from Leishmania donovani with 1,5-Bis (4-amidinophenoxy) pentane (PNT) at 2.95 A resolution
To Be Published
|
|
8IMR
 
 | | Structure of ligand-free human macrophage migration inhibitory factor | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor, ... | | Authors: | Sugishima, K, Noguchi, K, Yohda, M, Odaka, M, Matsumura, H. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Identification of methotrexate as an inhibitor of macrophage migration inhibitory factor by high-resolution crystal structure analysis
To Be Published
|
|
8IOO
 
 | |
6O78
 
 | | Crystal structure of Csm1-Csm4 cassette in complex with pppApApA | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, MANGANESE (II) ION, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6TVV
 
 | | Crystal structure of 3'-5' RecJ exonuclease from M. Jannaschii | | Descriptor: | GLYCEROL, MANGANESE (II) ION, MjaRecJ | | Authors: | De March, M, Medagli, B, Krastanova, I, Saha, I, Pisani, F, Onesti, S. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of 3'-5' RecJ exonuclease from M. Jannaschii
To Be Published
|
|
1TMZ
 
 | | TMZIP: A CHIMERIC PEPTIDE MODEL OF THE N-TERMINUS OF ALPHA TROPOMYOSIN, NMR, 15 STRUCTURES | | Descriptor: | TMZIP | | Authors: | Greenfield, N.J, Montelione, G.T, Hitchcock-Degregori, S.E, Farid, R.S. | | Deposit date: | 1998-04-20 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the N-terminus of striated muscle alpha-tropomyosin in a chimeric peptide: nuclear magnetic resonance structure and circular dichroism studies.
Biochemistry, 37, 1998
|
|
5BNN
 
 | | Crystal structure of human enterovirus D68 in complex with 6'SL | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
