2B98
 
 | | Crystal Structure of an archaeal pentameric riboflavin synthase | | Descriptor: | Riboflavin synthase | | Authors: | Ramsperger, A, Augustin, M, Schott, A.K, Gerhardt, S, Krojer, T, Eisenreich, W, Illarionov, B, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2005-10-11 | | Release date: | 2005-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an Archaeal Pentameric Riboflavin Synthase in Complex with a Substrate Analog Inhibitor: stereochemical implications
J.Biol.Chem., 281, 2006
|
|
3FUE
 
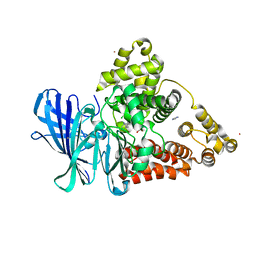 | |
5MWW
 
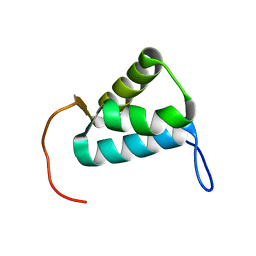 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
7ARV
 
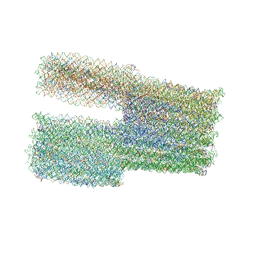 | | TwistTower_native-twist | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
7ARY
 
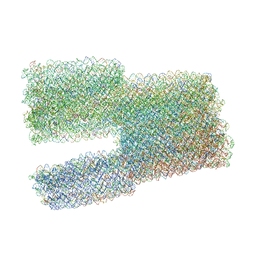 | | Twist-Tower_twist-corrected-variant | | Descriptor: | SCAFFOLD STRAND, STAPLE STRAND | | Authors: | Kube, M, Kohler, F, Feigl, E, Nagel-Yuksel, B, Willner, E.M, Funke, J.J, Gerling, T, Stommer, P, Honemann, M.N, Martin, T.G, Scheres, S.H.W, Dietz, H. | | Deposit date: | 2020-10-26 | | Release date: | 2020-11-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Revealing the structures of megadalton-scale DNA complexes with nucleotide resolution.
Nat Commun, 11, 2020
|
|
1YTI
 
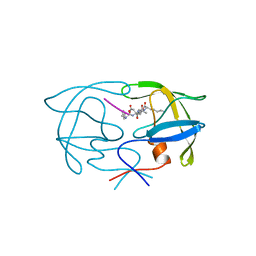 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | PEPTIDE PRODUCT, SIV PROTEASE | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
4BF1
 
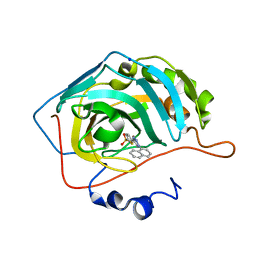 | | Three dimensional structure of human carbonic anhydrase II in complex with 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide | | Descriptor: | 5-(1-naphthalen-1-yl-1,2,3-triazol-4-yl)thiophene-2-sulfonamide, CARBONIC ANHYDRASE 2, SODIUM ION, ... | | Authors: | Tars, K, Leitans, J, Zalubovskis, R. | | Deposit date: | 2013-03-13 | | Release date: | 2014-01-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 5-Substituted-(1,2,3-Triazol-4-Yl)Thiophene-2-Sulfonamides Strongly Inhibit Human Carbonic Anhydrases I, II, Ix and Xii: Solution and X-Ray Crystallographic Studies.
Bioorg.Med.Chem., 21, 2013
|
|
5NCH
 
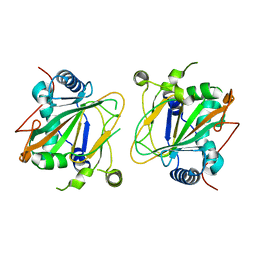 | | GriE apo form | | Descriptor: | Leucine hydroxylase | | Authors: | Lukat, P, Blankenfeldt, W, Mueller, R. | | Deposit date: | 2017-03-05 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Biosynthesis of methyl-proline containing griselimycins, natural products with anti-tuberculosis activity.
Chem Sci, 8, 2017
|
|
5UIM
 
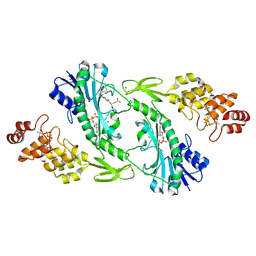 | | X-ray structure of the FdtF N-formyltransferase from salmonella enteric O60 in complex with folinic acid and TDP-Qui3N | | Descriptor: | Formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, POTASSIUM ION, ... | | Authors: | Woodford, C.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular architecture of an N-formyltransferase from Salmonella enterica O60.
J. Struct. Biol., 200, 2017
|
|
7B2W
 
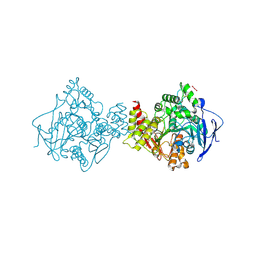 | | Torpedo californica acetylcholinesterase complexed with UO2 | | Descriptor: | Acetylcholinesterase, URANYL (VI) ION | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-28 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7XH2
 
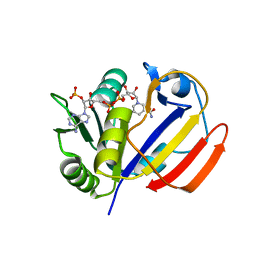 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
1WR4
 
 | |
7XH4
 
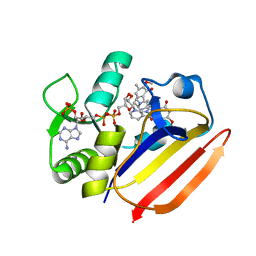 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis complex with safracin A | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Safracin A, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Zhang, Y, Yang, D, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
3BNH
 
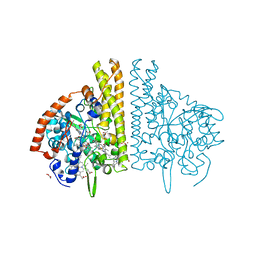 | | W. succinogenes NrfA Y218F Nitrite Complex | | Descriptor: | ACETATE ION, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Lukat, P, Einsle, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Binding and Reduction of Sulfite by Cytochrome c Nitrite Reductase
Biochemistry, 47, 2008
|
|
3HP2
 
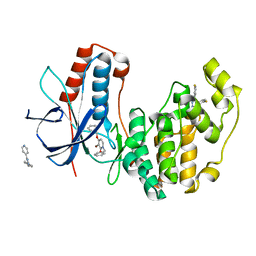 | | Crystal Structure of Human p38alpha complexed with a pyridinone compound | | Descriptor: | 1-benzyl-4-(benzyloxy)-3-bromopyridin-2(1H)-one, 2-fluoro-4-[4-(4-fluorophenyl)-1H-pyrazol-3-yl]pyridine, Mitogen-activated protein kinase 14 | | Authors: | Shieh, H.-S, Williams, J.M, Stegeman, R.A, Kurumbail, R.G. | | Deposit date: | 2009-06-03 | | Release date: | 2009-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of N-substituted pyridinones as potent and selective inhibitors of p38 kinase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6PC4
 
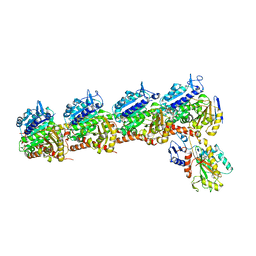 | | Tubulin-RB3_SLD-TTL in complex with compound ABI-274 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kumar, G, Wang, Y, Li, W, White, S.W. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structure-Activity Relationship Study of Novel 6-Aryl-2-benzoyl-pyridines as Tubulin Polymerization Inhibitors with Potent Antiproliferative Properties.
J.Med.Chem., 63, 2020
|
|
2PS6
 
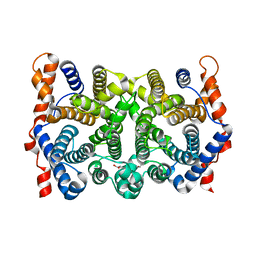 | | N225D/S229T trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, Trichodiene synthase | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-05-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif.
Arch.Biochem.Biophys., 469, 2008
|
|
2BGA
 
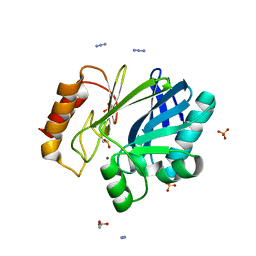 | | Bacillus cereus metallo-beta-lactamase (BcII) Arg (121) Cys mutant. Solved at pH7 using 20 Micromolar ZnSO4 in the buffer. 1mM DTT was used as a reducing agent. Cys221 is oxidized. | | Descriptor: | AZIDE ION, BETA-LACTAMASE II, GLYCEROL, ... | | Authors: | Davies, A.M, Rasia, R.M, Vila, A.J, Sutton, B.J, Fabiane, S.M. | | Deposit date: | 2004-12-17 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Effect of Ph on the Active Site of an Arg121Cys Mutant of the Metallo-Beta-Lactamase from Bacillus Cereus: Implications for the Enzyme Mechanism
Biochemistry, 44, 2005
|
|
3FUF
 
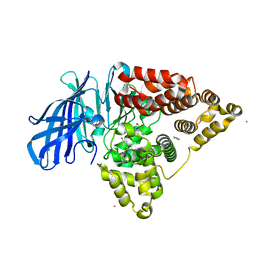 | |
3FU3
 
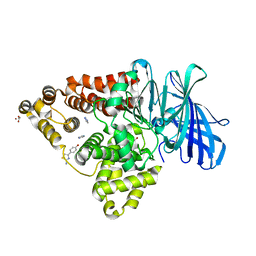 | |
3FUK
 
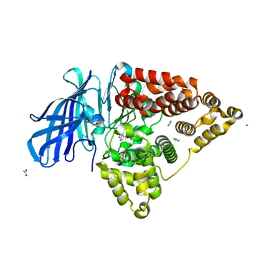 | |
8BIN
 
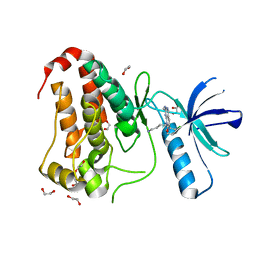 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with MR21 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-(2-chlorophenyl)-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Ephrin type-A receptor 2 | | Authors: | Zhubi, R, Rak, M, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Back-Pocket Optimization of 2-Aminopyrimidine-Based Macrocycles Leads to Potent EPHA2/GAK Kinase Inhibitors.
J.Med.Chem., 67, 2024
|
|
2PS7
 
 | | Y295F trichodiene synthase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Trichodiene synthase | | Authors: | Vedula, L.S, Cane, D.E, Christianson, D.W. | | Deposit date: | 2007-05-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and mechanistic analysis of trichodiene synthase using site-directed mutagenesis: probing the catalytic function of tyrosine-295 and the asparagine-225/serine-229/glutamate-233-Mg2+B motif.
Arch.Biochem.Biophys., 469, 2008
|
|
8DT9
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L141R with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8E5C
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L50F with Nirmatrelvir captured in two conformational states | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-08-20 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
