6T9B
 
 | | Crystal structrue of RSL W31A lectin mutant in complex with alpha-methylfucoside | | Descriptor: | Fucose-binding lectin protein, GLYCINE, methyl alpha-L-fucopyranoside | | Authors: | Houser, J, Komarek, J, Kozmon, S, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Chemistry, 26, 2020
|
|
7PUX
 
 | | Structure of p97 N-D1(L198W) in complex with Fragment TROLL2 | | Descriptor: | (1S)-2-amino-1-(4-bromophenyl)ethan-1-ol, ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bothe, S, Schindelin, H. | | Deposit date: | 2021-10-01 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Fragment screening using biolayer interferometry reveals ligands targeting the SHP-motif binding site of the AAA+ ATPase p97
Commun Chem, 5, 2022
|
|
7O07
 
 | |
7NC4
 
 | |
6T9A
 
 | | Crystal structrue of RSL W31FW76F lectin mutant in complex with L-fucose | | Descriptor: | 1,2-ETHANEDIOL, Fucose-binding lectin protein, alpha-L-fucopyranose, ... | | Authors: | Houser, J, Kozmon, S, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CH-pi Interaction in Protein-Carbohydrate Binding: Bioinformatics and In Vitro Quantification.
Chemistry, 26, 2020
|
|
7M2T
 
 | |
8GW0
 
 | | Crystal structure of the human dihydroorotase domain in complex with malic acid | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CAD protein, ZINC ION | | Authors: | Yang, P.C, Liu, H.W, Huang, H.Y, Huang, C.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil.
Biomolecules, 13, 2023
|
|
8GVZ
 
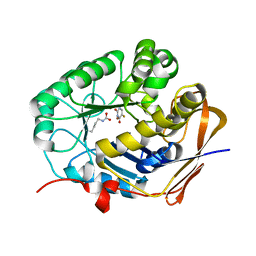 | | Crystal structure of the human dihydroorotase domain in complex with the anticancer drug 5-fluorouracil | | Descriptor: | 5-FLUOROURACIL, CAD protein, ZINC ION | | Authors: | Liu, H.W, Yang, P.C, Huang, H.Y, Huang, C.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Complexed Crystal Structure of the Dihydroorotase Domain of Human CAD Protein with the Anticancer Drug 5-Fluorouracil.
Biomolecules, 13, 2023
|
|
8GW5
 
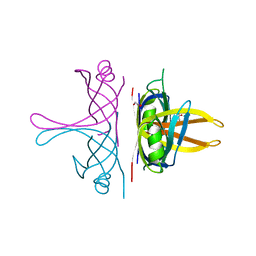 | |
5L88
 
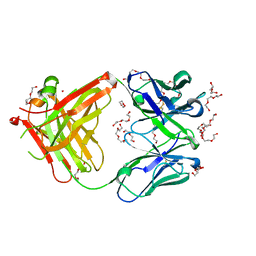 | | AFAMIN ANTIBODY FRAGMENT, N14 FAB, L1- GLYCOSILATED, CRYSTAL FORM I, non-parsimonious model | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Anti-afamin antibody N14, Fab fragment, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4OA8
 
 | |
5LGH
 
 | | Afamin antibody fragment, N14 Fab, L1- glycosilated, crystal form II, same as 5L7X, but isomorphous setting indexed same as 5L88, 5L9D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, MOUSE ANTIBODY FAB FRAGMENT, ... | | Authors: | Rupp, B, Naschberger, A. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The N14 anti-afamin antibody Fab: a rare VL1 CDR glycosylation, crystallographic re-sequencing, molecular plasticity and conservative versus enthusiastic modelling.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4MU6
 
 | |
7R97
 
 | | Crystal structure of postcleavge complex of Escherichia coli RNase III | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Dharavath, S, Shaw, G.X, Ji, X. | | Deposit date: | 2021-06-28 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Structural basis for Dicer-like function of an engineered RNase III variant and insights into the reaction trajectory of two-Mg 2+ -ion catalysis.
Rna Biol., 19, 2022
|
|
6Y9S
 
 | | Crystal structure of GSK-3b in complex with the imidazo[1,5-a]pyridine-3-carboxamide inhibitor 16 | | Descriptor: | ACETATE ION, Glycogen synthase kinase-3 beta, ~{N}-(oxan-4-ylmethyl)-6-(5-propan-2-yloxypyridin-3-yl)imidazo[1,5-a]pyridine-3-carboxamide | | Authors: | Krapp, S, Griessner, A, Blaesse, M, Buonfiglio, R, Ombrato, R. | | Deposit date: | 2020-03-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of Novel Imidazopyridine GSK-3 beta Inhibitors Supported by Computational Approaches.
Molecules, 25, 2020
|
|
5Q10
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N-[3-(acetylamino)phenyl]-4-chloro-N-[(1S)-1-cyclohexyl-2-(cyclohexylamino)-2-oxoethyl]benzamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q1D
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexyl-N-phenylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0J
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-[2-(5-phenylthiophen-2-yl)-1H-benzimidazol-1-yl]acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0X
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 6-(4-{[3-(3,5-dichloropyridin-4-yl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-2-methylphenyl)-1-methyl-1H-indole-3-carbox ylic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q15
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-N,2-dicyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0P
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 4-{(2S)-2-[2-(4-chlorophenyl)-5,6-difluoro-1H-benzimidazol-1-yl]-2-cyclohexylethoxy}benzoic acid, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q0T
 
 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | 2-phenyl-N-(propan-2-yl)-6-[(thiophen-2-yl)sulfonyl]-4,5,6,7-tetrahydro-1H-pyrrolo[2,3-c]pyridine-1-carboxamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q11
 
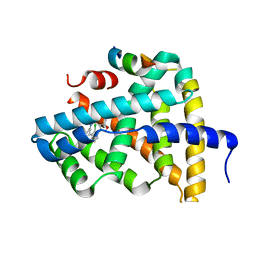 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3, N,N-dicyclohexyl-3-(2,4-dichlorophenyl)-5-methyl-1,2-oxazole-4-carboxamide | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q16
 
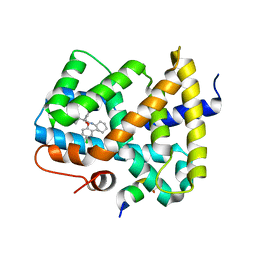 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-{2-[(4-chloro-2-methylphenoxy)methyl]-6-fluoro-1H-benzimidazol-1-yl}-N,2-dicyclohexylacetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
5Q18
 
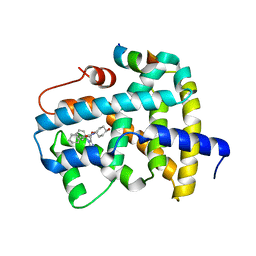 | | Ligand binding to FARNESOID-X-RECEPTOR | | Descriptor: | (2S)-2-cyclohexyl-2-{5,6-difluoro-2-[(R)-methoxy(phenyl)methyl]-1H-benzimidazol-1-yl}-N-(trans-4-hydroxycyclohexyl)acetamide, Bile acid receptor, COACTIVATOR PEPTIDE SRC-1 HD3 | | Authors: | Rudolph, M.G, Benz, J, Burger, D, Thoma, R, Ruf, A, Joseph, C, Kuhn, B, Shao, C, Yang, H, Burley, S.K. | | Deposit date: | 2017-05-31 | | Release date: | 2017-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D3R Grand Challenge 2: blind prediction of protein-ligand poses, affinity rankings, and relative binding free energies.
J. Comput. Aided Mol. Des., 32, 2018
|
|
