8SSF
 
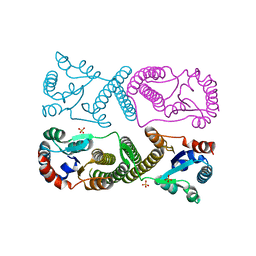 | | Minimal protein-only/RNA-free Ribonuclease P from Hydrogenobacter thermophilus | | Descriptor: | RNA-free ribonuclease P, SULFATE ION | | Authors: | Mendoza, J, Mallik, L, Wilhelm, C.A, Koutmos, M. | | Deposit date: | 2023-05-08 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Bacterial RNA-free RNase P: Structural and functional characterization of multiple oligomeric forms of a minimal protein-only ribonuclease P.
J.Biol.Chem., 299, 2023
|
|
4BBS
 
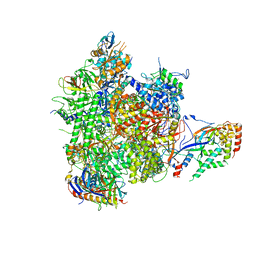 | | Structure of an initially transcribing RNA polymerase II-TFIIB complex | | Descriptor: | 5'-D(*AP*GP*CP*GP*CP*AP*GP*TP*TP*GP*TP*GP*CP*TP *AP*TP*GP*AP*TP*AP*TP*TP*TP*TP*TP*AP*TP)-3', 5'-D(*GP*GP*CP*AP*CP*AP*AP*CP*TP*GP*CP*GP*CP*TP)-3', 5'-R(*AP*UP*AP*UP*CP*AP)-3', ... | | Authors: | Sainsbury, S, Niesser, J, Cramer, P. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure and Function of the Initially Transcribing RNA Polymerase II-TFIIB Complex
Nature, 493, 2013
|
|
5LTJ
 
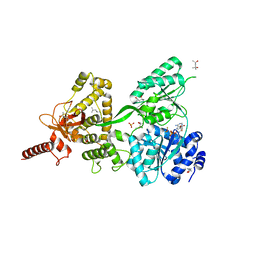 | | Crystal structure of the Prp43-ADP-BeF3 complex (in orthorhombic space group) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tauchert, M.J, Ficner, R. | | Deposit date: | 2016-09-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural insights into the mechanism of the DEAH-box RNA helicase Prp43.
Elife, 6, 2017
|
|
1B75
 
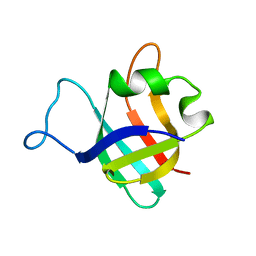 | |
8SSG
 
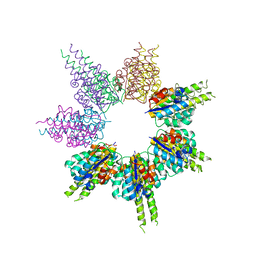 | |
5LTK
 
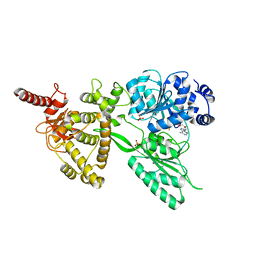 | |
2KM8
 
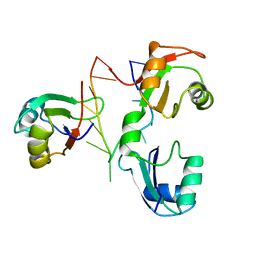 | |
3HOW
 
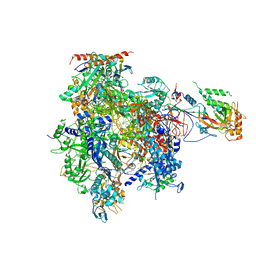 | | Complete RNA polymerase II elongation complex III with a T-U mismatch and a frayed RNA 3'-uridine | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
8UTJ
 
 | |
8UXB
 
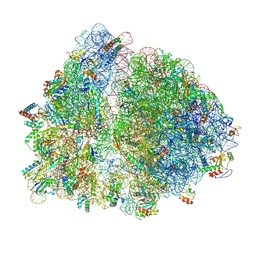 | |
8UZ3
 
 | |
1RC7
 
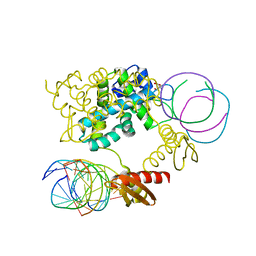 | | Crystal structure of RNase III Mutant E110K from Aquifex Aeolicus complexed with ds-RNA at 2.15 Angstrom Resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*GP*GP*CP*GP*CP*GP*CP*GP*CP*C)-3', Ribonuclease III | | Authors: | Blaszczyk, J, Gan, J, Ji, X. | | Deposit date: | 2003-11-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Noncatalytic Assembly of Ribonuclease III with Double-Stranded RNA.
Structure, 12, 2004
|
|
8UZG
 
 | |
3HOU
 
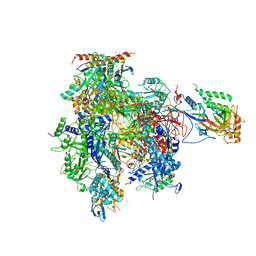 | | Complete RNA polymerase II elongation complex I with a T-U mismatch | | Descriptor: | 5'-D(*A*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*CP*A*AP*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*U*UP*CP*GP*AP*CP*CP*AP*GP*GP*CP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
3HOZ
 
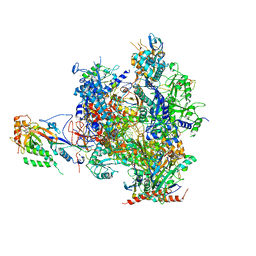 | | Complete RNA polymerase II elongation complex IV with a T-U mismatch and a frayed RNA 3'-guanine | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*CP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*G)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
7OG5
 
 | |
5M9Z
 
 | | Second zinc-binding domain from yeast Pcf11 | | Descriptor: | Protein PCF11, ZINC ION | | Authors: | Mackereth, C. | | Deposit date: | 2016-11-02 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Distinct roles of Pcf11 zinc-binding domains in pre-mRNA 3'-end processing.
Nucleic Acids Res., 45, 2017
|
|
6JVX
 
 | | Crystal structure of RBM38 in complex with RNA | | Descriptor: | RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*GP*UP*GP*UP*G)-3'), RNA-binding protein 38, SULFATE ION | | Authors: | Qian, K, Li, M, Wang, J, Zhang, M, Wang, M. | | Deposit date: | 2019-04-17 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis for mRNA recognition by human RBM38.
Biochem.J., 477, 2020
|
|
3HOY
 
 | | Complete RNA polymerase II elongation complex VI | | Descriptor: | 5'-D(*CP*CP*AP*AP*GP*CP*TP*CP*AP*AP*G*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*TP*AP*CP*TP*AP*GP*TP*AP*CP*TP*GP*CP*C)-3', 5'-D(*CP*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*AP*GP*TP*AP*AP*AP*CP*TP*AP*GP*TP*AP*TP*T*GP*AP*AP*AP*GP*TP*AP*CP*TP*TP*GP*AP*GP*CP*TP*T)-3', 5'-R(*UP*AP*UP*AP*UP*GP*CP*A*UP*AP*AP*AP*GP*AP*CP*CP*AP*GP*GP*A)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
3HOX
 
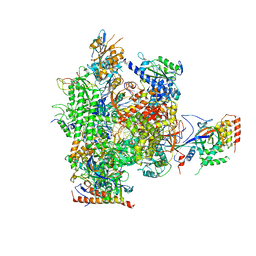 | | Complete RNA polymerase II elongation complex V | | Descriptor: | 5'-D(*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*C*AP*AP*GP*TP*AP*GP*TP*TP*AP*AP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*U*CP*AP*AP*CP*CP*AP*GP*GP*CP*UP*U)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
6QPG
 
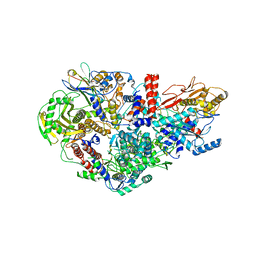 | | Influenza A virus Polymerase Heterotrimer A/nt/60/1968(H3N2) in complex with Nanobody NB8205 | | Descriptor: | Nanobody NB8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Fan, H.T, Keown, J.R, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-02-13 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.34 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
3HOV
 
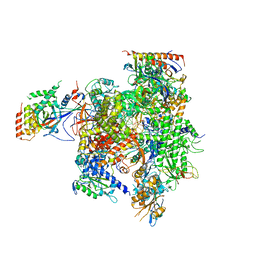 | | Complete RNA polymerase II elongation complex II | | Descriptor: | 5'-D(*AP*GP*CP*TP*CP*AP*A*GP*TP*AP*GP*TP*TP*AP*TP*GP*CP*CP*(BRU)P*GP*GP*TP*CP*AP*TP*T)-3', 5'-D(*T*AP*CP*TP*AP*CP*TP*TP*GP*AP*GP*CP*T)-3', 5'-R(*UP*GP*CP*AP*UP*UP*UP*CP*GP*AP*CP*CP*AP*GP*GP*CP*A)-3', ... | | Authors: | Sydow, J.F, Brueckner, F, Cheung, A.C.M, Damsma, G.E, Dengl, S, Lehmann, E, Vassylyev, D, Cramer, P. | | Deposit date: | 2009-06-03 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: mismatch-specific fidelity mechanisms and paused RNA polymerase II with frayed RNA.
Mol.Cell, 34, 2009
|
|
6KUR
 
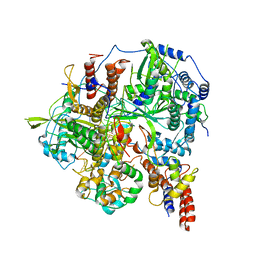 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
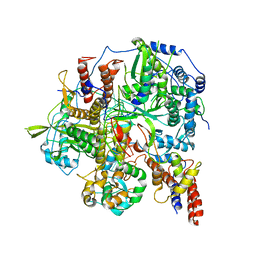 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUP
 
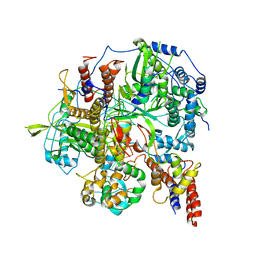 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
