6UTB
 
 | |
8UXN
 
 | | Caulobacter crescentus FljM flagellar filament (symmetrized) | | Descriptor: | Flagellin FljM | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2023-11-09 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
6USW
 
 | | CRYSTAL STRUCTURE OF HIV-1 LM/HS CLADE A/E CRF01 GP120 CORE IN COMPLEX WITH (S)-MCG-IV-210 | | Descriptor: | (3S)-N~1~-(2-aminoethyl)-N~3~-(4-chloro-3-fluorophenyl)piperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-10-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HIV-1 Env gp120 Inner Domain Shapes the Phe43 Cavity and the CD4 Binding Site.
Mbio, 11, 2020
|
|
3DWA
 
 | | Crystal structure of the B-subunit of the AB5 toxin from E. coli | | Descriptor: | PENTAETHYLENE GLYCOL, Subtilase cytotoxin, subunit B | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.084 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
3DWP
 
 | | Crystal structure of the B-subunit of the AB5 toxin from E. Coli with Neu5Gc | | Descriptor: | N-glycolyl-alpha-neuraminic acid, PENTAETHYLENE GLYCOL, Subtilase cytotoxin, ... | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
7PWO
 
 | | Cryo-EM structure of Giardia lamblia ribosome at 2.75 A resolution | | Descriptor: | 40S ribosomal protein S21, 40S ribosomal protein S26, 40S ribosomal protein S30, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-07 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
7QCR
 
 | | MLLT4/Afadin PDZ domain in complex with the C-terminal peptide from protein E of SARS-CoV-2 | | Descriptor: | Afadin, Envelope small membrane protein, SULFATE ION | | Authors: | Zhu, Y, Alvarez, F, Haouz, A, Mechaly, A, Caillet-Saguy, C. | | Deposit date: | 2021-11-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Interactions of Severe Acute Respiratory Syndrome Coronavirus 2 Protein E With Cell Junctions and Polarity PSD-95/Dlg/ZO-1-Containing Proteins.
Front Microbiol, 13, 2022
|
|
7PWG
 
 | | Cryo-EM structure of large subunit of Giardia lamblia ribosome at 2.7 A resolution | | Descriptor: | 60S ribosomal protein L13, 60S ribosomal protein L18a, 60S ribosomal protein L27, ... | | Authors: | Hiregange, D.G, Rivalta, A, Bose, T, Breiner-Goldstein, E, Samiya, S, Cimicata, G, Kulakova, L, Zimmerman, E, Bashan, A, Herzberg, O, Yonath, A. | | Deposit date: | 2021-10-06 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structure of the ancient eukaryotic ribosome from the human parasite Giardia lamblia.
Nucleic Acids Res., 50, 2022
|
|
3E8Z
 
 | |
6N62
 
 | | Escherichia coli RNA polymerase sigma70-holoenzyme bound to upstream fork promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Braffman, N, Hauver, J, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-24 | | Release date: | 2019-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.803 Å) | | Cite: | Structural mechanism of transcription inhibition by lasso peptides microcin J25 and capistruin.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8UXK
 
 | | Caulobacter crescentus FljL flagellar filament (asymmetrical) | | Descriptor: | Flagellin | | Authors: | Sanchez, J.C, Montemayor, E.J, Ploscariu, N.T, Parrell, D, Baumgardt, J.K, Yang, J.E, Sibert, B, Cai, K, Wright, E.R. | | Deposit date: | 2023-11-09 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct evidence for multi-flagellin filament stabilization via atomic-level architecture of Caulobacter crescentus flagellar filaments
To Be Published
|
|
7A3P
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Single Chain Variable Fragment | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3Q
 
 | | Crystal structure of dengue 4 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7OQE
 
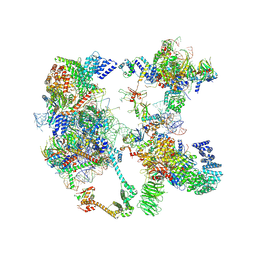 | | Saccharomyces cerevisiae spliceosomal pre-A complex (delta BS-A ACT1) | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, ACT1 pre-mRNA (delta BS-A), Cold sensitive U2 snRNA suppressor 1, ... | | Authors: | Zhang, Z, Rigo, N, Dybkov, O, Fourmann, J, Will, C.L, Kumar, V, Urlaub, H, Stark, H, Luehrmann, R. | | Deposit date: | 2021-06-03 | | Release date: | 2021-08-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural insights into how Prp5 proofreads the pre-mRNA branch site.
Nature, 596, 2021
|
|
3E9B
 
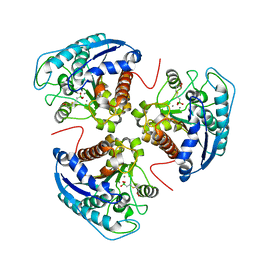 | |
7ZB6
 
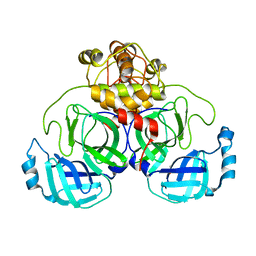 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) variant C44S at 2.12 A resolution | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Paknia, E, Rabe von Pappenheim, F, Funk, L.-M, Tittmann, K, Chari, A. | | Deposit date: | 2022-03-23 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Multiple redox switches of the SARS-CoV-2 main protease in vitro provide opportunities for drug design.
Nat Commun, 15, 2024
|
|
8SBQ
 
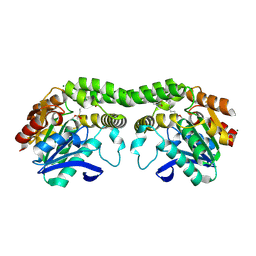 | |
8PVW
 
 | |
8BU6
 
 | | Structure of DDB1 bound to DS55-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUK
 
 | | Structure of DDB1 bound to DS08-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-(naphthalen-2-ylmethylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU2
 
 | | Structure of DDB1 bound to DS18-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUI
 
 | | Structure of DDB1 bound to DRF-053-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[9-propan-2-yl-6-[(4-pyridin-2-ylphenyl)amino]purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU3
 
 | | Structure of DDB1 bound to DS19-engaged CDK12-cyclin K | | Descriptor: | 1,2-ETHANEDIOL, 2-morpholin-4-yl-9-propan-2-yl-~{N}-[(4-pyridin-2-ylphenyl)methyl]purin-6-amine, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUL
 
 | | Structure of DDB1 bound to DS11-engaged CDK12-cyclin K | | Descriptor: | (2~{R})-2-[[6-(3-phenylpropylamino)-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, 1,2-ETHANEDIOL, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BU5
 
 | | Structure of DDB1 bound to SR-4835-engaged CDK12-cyclin K | | Descriptor: | CITRIC ACID, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.134 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
