7FY6
 
 | | Crystal Structure of human FABP4 in complex with 6-phenyl-11H-pyrimido[4,5-c][2]benzazepin-3-amine, i.e. SMILES C1(=Nc2c(Cc3c1cccc3)cnc(n2)N)c1ccccc1 with IC50=1.4 microM | | Descriptor: | (10S)-10-phenyl-10,11-dihydro-5H-pyrimido[4,5-c][2]benzazepin-2-amine, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Fryer, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
4UJD
 
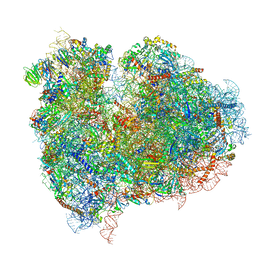 | | mammalian 80S HCV-IRES initiation complex with eIF5B PRE-like state | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S RIBOSOMAL PROTEIN ES1, ... | | Authors: | Yamamoto, H, Unbehaun, A, Loerke, J, Behrmann, E, Marianne, C, Burger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the Mammalian 80S Initiation Complex with Eif5B on Hcv Ires
Nat.Struct.Mol.Biol., 21, 2014
|
|
5QCJ
 
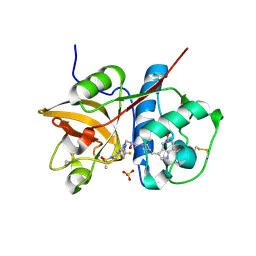 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 5-hydroxy-3-{1-[(2S)-2-hydroxy-3-{5-(methylsulfonyl)-3-[4-(trifluoromethyl)phenyl]-4,5,6,7-tetrahydro-1H-pyrazolo[4,3-c]pyridin-1-yl}propyl]piperidin-4-yl}-1H-pyrrolo[3,2-c]pyridin-5-ium, Cathepsin S, SULFATE ION | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
5QCH
 
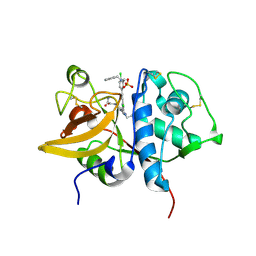 | | Crystal structure of human Cathepsin-S with bound ligand | | Descriptor: | 2-(3-[3-({3-[(benzylamino)methyl]-4-chlorophenyl}ethynyl)-4-chlorophenyl]-1-{3-[(3S)-3-methylmorpholin-4-yl]propyl}-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)-2-oxoacetamide, Cathepsin S, GLYCEROL, ... | | Authors: | Bembenek, S.D, Ameriks, M.K, Mirzadegan, T, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-08-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human Cathepsin-S with bound ligand
To be published
|
|
7FWZ
 
 | | Crystal Structure of human FABP4 in complex with 4-[3-(trifluoromethyl)-5,6,7,8-tetrahydro-4H-cyclohepta[c]pyrazol-1-yl]butanoic acid, i.e. SMILES C1(=NN(C2=C1CCCCC2)CCCC(=O)O)C(F)(F)F with IC50=0.109 microM | | Descriptor: | 4-[3-(trifluoromethyl)-5,6,7,8-tetrahydrocyclohepta[c]pyrazol-1(4H)-yl]butanoic acid, FORMIC ACID, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal Structure of a human FABP4 complex
To be published
|
|
7F2D
 
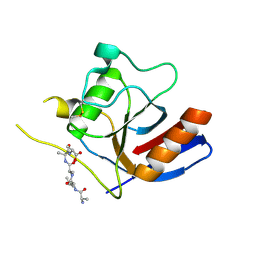 | |
7F2I
 
 | |
6LAE
 
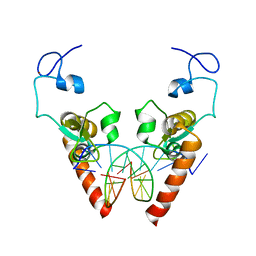 | | Crystal structure of the DNA-binding domain of human XPA in complex with DNA | | Descriptor: | DNA (5'-D(P*GP*CP*AP*TP*CP*TP*CP*GP*CP*CP*T)-3'), DNA (5'-D(P*TP*GP*GP*CP*GP*AP*GP*AP*TP*GP*C)-3'), DNA repair protein complementing XP-A cells, ... | | Authors: | Lian, F.M, Yang, X, Jiang, Y.L, Yang, F, Li, C, Yang, W, Qian, C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | New structural insights into the recognition of undamaged splayed-arm DNA with a single pair of non-complementary nucleotides by human nucleotide excision repair protein XPA.
Int.J.Biol.Macromol., 148, 2020
|
|
7FV8
 
 | | PanDDA analysis group deposition -- PHIP in complex with Z964297186 | | Descriptor: | 4-(3-chlorobenzoyl)-N-[3-(6,7-dihydrothieno[3,2-c]pyridin-5(4H)-yl)-3-oxopropyl]-1,4-diazepane-1-carboxamide, PH-interacting protein | | Authors: | Grosjean, H, Tomlinson, C, Bradshaw, W.J, Koekemoer, L, Krojer, T, Fearon, D, Biggin, P.C, von Delft, F. | | Deposit date: | 2023-03-09 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
9MOT
 
 | | Cryo-EM structure of factor Va bound to activated protein C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor Va heavy chain, Coagulation factor Va light chain, ... | | Authors: | Mohammed, B.M, Basore, K, Di Cera, E. | | Deposit date: | 2024-12-27 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structure of coagulation factor Va bound to activated protein C.
Blood, 145, 2025
|
|
9ODS
 
 | | Structure of CRBN TBD bound to compound C3 | | Descriptor: | (1P)-1-[(4S)-8H-spiro[furo[2,3-c]imidazo[1,2-a]pyridine-7,4'-piperidin]-3-yl]pyrimidine-2,4(1H,3H)-dione, Protein cereblon, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2025-04-27 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
9MOV
 
 | | Cryo-EM structure of factor Va bound to activated protein C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Coagulation factor Va heavy chain, ... | | Authors: | Mohammed, B.M, Basore, K, Di Cera, E. | | Deposit date: | 2024-12-27 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of coagulation factor Va bound to activated protein C.
Blood, 145, 2025
|
|
6LXN
 
 | |
9M56
 
 | | Bovine Heart Cytochrome c Oxidase in the Fully Reduced State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2025-03-05 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 2025
|
|
7FEA
 
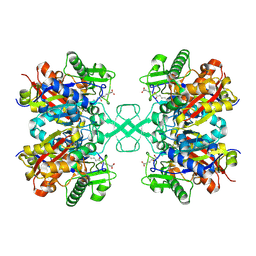 | | PY14 in complex with Col-D | | Descriptor: | (6~{R},7~{R},9~{E})-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Ko, T.P, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
7FVS
 
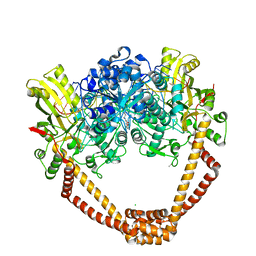 | | Crystal Structure of S. aureus gyrase in complex with 4-[[1-[(1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl)methyl]azetidin-3-yl]methylamino]-6-fluorochromen-2-one | | Descriptor: | 4-{[(1-{[(6R)-1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl]methyl}azetidin-3-yl)methyl]amino}-6-fluoro-2H-1-benzopyran-2-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Xu, B, Benz, J, Cumming, J.G, Kreis, L, Rudolph, M.G. | | Deposit date: | 2023-04-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
7OJJ
 
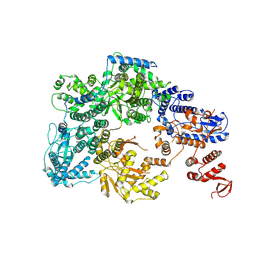 | | Lassa virus L protein with endonuclease and C-terminal domains in close proximity [MID-LINK] | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7O9U
 
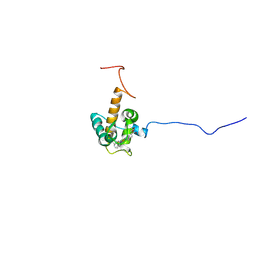 | | Solution structure of oxidized cytochrome c552 from Thioalkalivibrio paradoxus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Britikov, V.V, Britikova, E.V, Altukhov, D.A, Timofeev, V.I, Dergousova, N.I, Rakitina, T.V, Tikhonova, T.V, Usanov, S.A, Popov, V.O, Bocharov, E.V. | | Deposit date: | 2021-04-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Unusual Cytochrome c 552 from Thioalkalivibrio paradoxus : Solution NMR Structure and Interaction with Thiocyanate Dehydrogenase.
Int J Mol Sci, 23, 2022
|
|
6OD2
 
 | | Yeast Spc42 C-terminal Antiparallel Coiled-Coil | | Descriptor: | Spindle pole body component SPC42 | | Authors: | Drennan, A.C, Krishna, S, Seeger, M.A, Andreas, M.P, Gardner, J.M, Sether, E.K.R, Jaspersen, S.L, Rayment, I. | | Deposit date: | 2019-03-25 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.435 Å) | | Cite: | Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication.
Mol.Biol.Cell, 30, 2019
|
|
7O6L
 
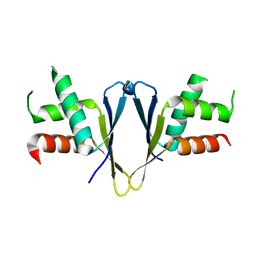 | | Crystal structure of C. elegans ERH-2 | | Descriptor: | Enhancer of rudimentary homolog 2 | | Authors: | Falk, S, Ketting, R.F. | | Deposit date: | 2021-04-11 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of PETISCO complex assembly during piRNA biogenesis in C. elegans .
Genes Dev., 35, 2021
|
|
7RQF
 
 | |
7RQH
 
 | |
7RPQ
 
 | |
8JB9
 
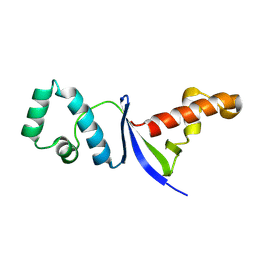 | | Solution structure of Anti-CRISPR protein AcrIIC5 | | Descriptor: | Type II-C anti-CRISPR protein, AcrIIC5 | | Authors: | Hong, S.H, Park, C, An, S.Y, Suh, J.Y. | | Deposit date: | 2023-05-08 | | Release date: | 2024-05-08 | | Last modified: | 2025-05-21 | | Method: | SOLUTION NMR | | Cite: | Structural variants of AcrIIC5 inhibit Cas9 via divergent binding interfaces.
Structure, 33, 2025
|
|
6GDL
 
 | |
