4YFX
 
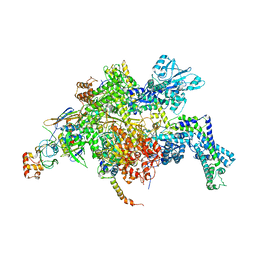 | | Escherichia coli RNA polymerase in complex with Myxopyronin B | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.844 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
3FNV
 
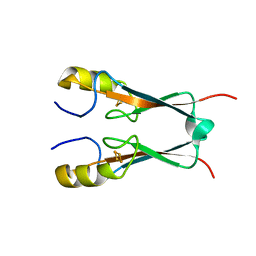 | | Crystal Structure of Miner1: The Redox-active 2Fe-2S Protein Causative in Wolfram Syndrome 2 | | Descriptor: | CDGSH iron sulfur domain-containing protein 2, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Conlan, A.R, Axelrod, H.L, Cohen, A.E, Abresch, E.C, Yee, D, Zuris, J, Nechushtai, R, Jennings, P.A, Paddock, M.L. | | Deposit date: | 2008-12-26 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of Miner1: The redox-active 2Fe-2S protein causative in Wolfram Syndrome 2.
J.Mol.Biol., 392, 2009
|
|
3F64
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-O)paranitrophenyl ligand | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, F17a-G | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-05 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
6O44
 
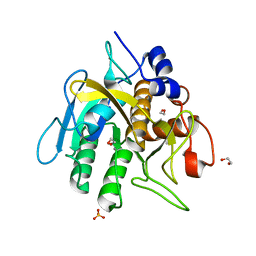 | | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Nattokinase, ... | | Authors: | Tang, H, Shi, K, Aihara, H. | | Deposit date: | 2019-02-28 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Insight into subtilisin E-S7 cleavage pattern based on crystal structure and hydrolysates peptide analysis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
9CJK
 
 | | Human TMED9 octamer structure | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
9CJL
 
 | | Molecular basis of TMED9 dodecamer | | Descriptor: | Transmembrane emp24 domain-containing protein 9, [(2R)-2-[(E)-octadec-9-enoyl]oxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] (E)-octadec-9-enoate | | Authors: | Le, X, Xiong, P. | | Deposit date: | 2024-07-06 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular basis of TMED9 oligomerization and entrapment of misfolded protein cargo in the early secretory pathway.
Sci Adv, 10, 2024
|
|
6OT6
 
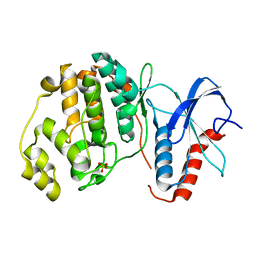 | | Rat ERK2 D319N | | Descriptor: | Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Taylor, C.A, Goldsmith, E.J, Cobb, M.H. | | Deposit date: | 2019-05-02 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional divergence caused by mutations in an energetic hotspot in ERK2.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4YFK
 
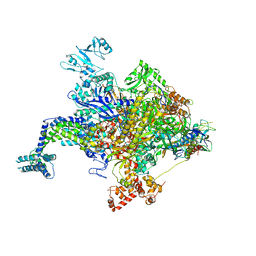 | | Escherichia coli RNA polymerase in complex with squaramide compound 8. | | Descriptor: | 3,5-dimethyl-N-{2-[4-(4-methylbenzyl)piperidin-1-yl]-3,4-dioxocyclobut-1-en-1-yl}-1,2-oxazole-4-sulfonamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.571 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4X9E
 
 | | DEOXYGUANOSINETRIPHOSPHATE TRIPHOSPHOHYDROLASE from Escherichia coli with two DNA effector molecules | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, MAGNESIUM ION, RNA (5'-R(P*CP*CP*C)-3') | | Authors: | Singh, D, Gawel, D, Itsko, M, Krahn, J.M, London, R.E, Schaaper, R.M. | | Deposit date: | 2014-12-11 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES.
J.Biol.Chem., 290, 2015
|
|
8EIZ
 
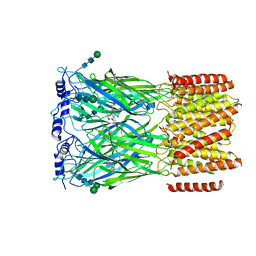 | | Cryo-EM structure of squid sensory receptor CRB1 | | Descriptor: | N-benzyl-2-(2,6-dimethylanilino)-N,N-diethyl-2-oxoethan-1-aminium, Squid sensory receptor CRB1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kang, G, Kim, J.J, Allard, C.A.H, Valencia-Montoya, W.A, van Giesen, L, Kilian, P.B, Bai, X, Bellono, N.W, Hibbs, R.E. | | Deposit date: | 2022-09-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Sensory specializations drive octopus and squid behaviour.
Nature, 616, 2023
|
|
6NCO
 
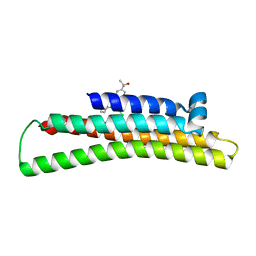 | | Fragment-based Discovery of an apoE4 Stabilizer | | Descriptor: | 1-[5-chloro-4'-(2-hydroxypropan-2-yl)[1,1'-biphenyl]-3-yl]cyclobutane-1-carboximidamide, Apolipoprotein E | | Authors: | Jakob, C.G, Qiu, W. | | Deposit date: | 2018-12-11 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.707 Å) | | Cite: | Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
J.Med.Chem., 62, 2019
|
|
4YFN
 
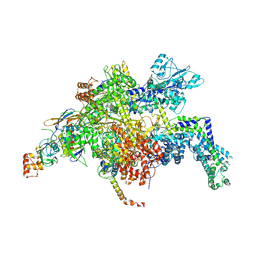 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
3FF8
 
 | | Structure of NK cell receptor KLRG1 bound to E-cadherin | | Descriptor: | CALCIUM ION, Epithelial cadherin, Killer cell lectin-like receptor subfamily G member 1 | | Authors: | Li, Y, Mariuzza, R.A. | | Deposit date: | 2008-12-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition.
Immunity, 31, 2009
|
|
3FFO
 
 | | F17b-G lectin domain with bound GlcNAc(beta1-2)man | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose, Adhesin, ... | | Authors: | Buts, L, De Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-12-04 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli.
Biology (Basel), 2, 2013
|
|
3F6J
 
 | | F17a-G lectin domain with bound GlcNAc(beta1-3)Gal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-methyl beta-D-galactopyranoside, F17a-G | | Authors: | Buts, L, de Boer, A, Olsson, J.D.M, Jonckheere, W, De Kerpel, M, De Genst, E, Guerardel, Y, Willaert, R, Wyns, L, Wuhrer, M, Oscarson, S, De Greve, H, Bouckaert, J. | | Deposit date: | 2008-11-06 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Sampling of Glycan Interaction Profiles Reveals Mucosal Receptors for Fimbrial Adhesins of Enterotoxigenic Escherichia coli
Biology, 2, 2013
|
|
5IWM
 
 | | 2.5A structure of GSK945237 with S.aureus DNA gyrase and DNA. | | Descriptor: | (1R)-1-[(4-{[(6,7-dihydro[1,4]dioxino[2,3-c]pyridazin-3-yl)methyl]amino}piperidin-1-yl)methyl]-9-fluoro-1,2-dihydro-4H-pyrrolo[3,2,1-ij]quinolin-4-one, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*TP*TP*CP*AP*CP*CP*GP*CP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*GP*CP*GP*GP*TP*GP*AP*AP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Bax, B.D, Miles, T.J. | | Deposit date: | 2016-03-22 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclics (e.g., GSK945237) as potent inhibitors of bacterial type IIA topoisomerases.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7TZK
 
 | | EPS8 SH3 Domain with NleH1 PxxDY Motif | | Descriptor: | Epidermal growth factor receptor kinase substrate 8, T3SS secreted effector NleH homolog | | Authors: | Grishin, A.M, Cygler, M. | | Deposit date: | 2022-02-15 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8UF6
 
 | | Structure of Trek-1(K2P2.1) with ML336 | | Descriptor: | CADMIUM ION, DECANE, DODECANE, ... | | Authors: | Lolicato, M, Mondal, A, Minor, D.L. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-26 | | Last modified: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of covalent chemogenetic K 2P channel activators.
Cell Chem Biol, 31, 2024
|
|
6Z1U
 
 | | bovine ATP synthase F1c8-peripheral stalk domain, state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F(0) complex subunit B1, ... | | Authors: | Spikes, T, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-05-14 | | Release date: | 2020-09-09 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7RWA
 
 | | AP2 bound to heparin and Tgn38 tyrosine cargo peptide | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6QI4
 
 | | NCS-1 bound to a ligand | | Descriptor: | 2-(1~{H}-indol-3-yl)-~{N}-[(~{E})-(4-nitro-3-oxidanyl-phenyl)methylideneamino]ethanamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Sanchez-Barrena, M.J, Blanco-Gabella, P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Insights into real-time chemical processes in a calcium sensor protein-directed dynamic library.
Nat Commun, 10, 2019
|
|
7RWB
 
 | | AP2 bound to the APA domain of SGIP in the presence of heparin | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8IZE
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A1 in complex with 4-HMBPP | | Descriptor: | Butyrophilin subfamily 3 member A1, DI(HYDROXYETHYL)ETHER, [(E)-3-(hydroxymethyl)pent-2-enyl] phosphono hydrogen phosphate | | Authors: | Yang, Y.Y, Yi, S.M, Huang, J.W, Chen, C.C, Guo, R.T. | | Deposit date: | 2023-04-07 | | Release date: | 2023-09-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Phosphoantigens glue butyrophilin 3A1 and 2A1 to activate V gamma 9V delta 2 T cells.
Nature, 621, 2023
|
|
7RWC
 
 | | AP2 bound to the APA domain of SGIP and heparin; partial signal subtraction and symmetry expansion | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Baker, R.W, Hollopeter, G, Partlow, E.A. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6U25
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS- RORGT(244-487)-L6-SRC1(678-692)) IN COMPLEX WITH A TRICYCLIC INVERSE AGONIST | | Descriptor: | GLYCEROL, NUCLEAR RECEPTOR COACTIVATOR 1 CHIMERA, trans-4-[(3aR,9bR)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl]cyclohexane-1-carboxylic acid | | Authors: | Sack, J. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Rationally Designed, Conformationally Constrained Inverse Agonists of ROR gamma t-Identification of a Potent, Selective Series with Biologic-Like in Vivo Efficacy.
J.Med.Chem., 62, 2019
|
|
