7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2R9Z
 
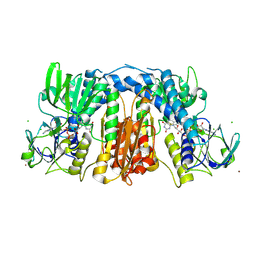 | | Glutathione amide reductase from Chromatium gracile | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione amide reductase, ... | | Authors: | Van Petegem, F, Vergauwen, B, Savvides, S, De Vos, D, Van Beeumen, J. | | Deposit date: | 2007-09-14 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding nicotinamide dinucleotide cofactor and substrate specificity in class I flavoprotein disulfide oxidoreductases: crystallographic analysis of a glutathione amide reductase.
J.Mol.Biol., 374, 2007
|
|
6IIH
 
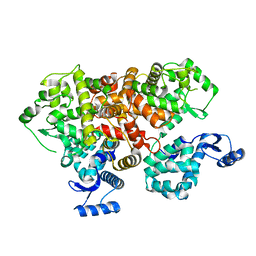 | | crystal structure of mitochondrial calcium uptake 2(MICU2) | | Descriptor: | CALCIUM ION, Endolysin,Calcium uptake protein 2, mitochondrial | | Authors: | Shen, Q, Wu, W, Zheng, J, Jia, Z. | | Deposit date: | 2018-10-06 | | Release date: | 2019-08-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | The crystal structure of MICU2 provides insight into Ca2+binding and MICU1-MICU2 heterodimer formation.
Embo Rep., 20, 2019
|
|
2RB1
 
 | |
6A36
 
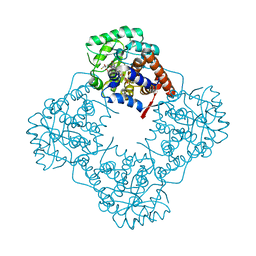 | | Mandelate oxidase mutant-Y128F with the 3-fluoropyruvic acid FMN adduct | | Descriptor: | 1-deoxy-1-{5-[(1S)-2-fluoro-1-hydroxyethyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-5-O-phosphono-D-ribitol, 1-{5-[(3S)-3-carboxy-4-fluoro-3-hydroxybutanoyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-1-deoxy-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
7Q2A
 
 | | Crystal structure of AphC in complex with 4-ethylcatechol | | Descriptor: | 4-ethylbenzene-1,2-diol, CALCIUM ION, Catechol 2,3-dioxygenase, ... | | Authors: | Zahn, M, Grigg, J.C, Eltis, L.D, McGeehan, J.E. | | Deposit date: | 2021-10-25 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Characterization of a phylogenetically distinct extradiol dioxygenase involved in the bacterial catabolism of lignin-derived aromatic compounds.
J.Biol.Chem., 298, 2022
|
|
5ZZR
 
 | | The crystal structure of Mandelate oxidase with (S)-mandelic acid | | Descriptor: | (S)-MANDELIC ACID, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-04 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | The flavin mononucleotide cofactor in alpha-hydroxyacid oxidases exerts its electrophilic/nucleophilic duality in control of the substrate-oxidation level.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
5ZZX
 
 | |
6A41
 
 | | Dehalogenation enzyme | | Descriptor: | dehalogenase | | Authors: | Yin, B, Yuan, A.Y. | | Deposit date: | 2018-06-18 | | Release date: | 2019-06-19 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of a new dehalogenase at 1.9 Angstroms resolution
To Be Published
|
|
6A00
 
 | | The crystal structure of Mandelate oxidase with (S)-2-phenylpropionate | | Descriptor: | (2~{S})-2-phenylpropanoic acid, 4-hydroxymandelate oxidase, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Biochemical and structural explorations of alpha-hydroxyacid oxidases reveal a four-electron oxidative decarboxylation reaction.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4LN1
 
 | | CRYSTAL STRUCTURE OF L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452 | | Descriptor: | CALCIUM ION, L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452
To be Published
|
|
3HF2
 
 | | Crystal structure of the I401P mutant of cytochrome P450 BM3 | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2009-05-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Highly Active Single-Mutation Variant of P450(BM3) (CYP102A1)
Chembiochem, 10, 2009
|
|
6IH8
 
 | |
2RDB
 
 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase I100W Mutant | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Murray, L.J, Garcia-Serres, R, McCormick, M.S, Davydov, R, Naik, S, Hoffman, B.M, Huynh, B.H, Lippard, S.J. | | Deposit date: | 2007-09-21 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dioxygen activation at non-heme diiron centers: oxidation of a proximal residue in the I100W variant of toluene/o-xylene monooxygenase hydroxylase.
Biochemistry, 46, 2007
|
|
7PVJ
 
 | | Crystal structure of Thioredoxin Reductase from Brugia Malayi in complex with auranofin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Ippoliti, R, Gencheva, R, Cheng, Q, Arner, E.S.J, Angelucci, F, Williams, D.L. | | Deposit date: | 2021-10-04 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
6A18
 
 | | Crystal structure of CYP90B1 in complex with 1,6-hexandiol | | Descriptor: | CHLORIDE ION, Cytochrome P450 90B1, GLYCEROL, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
3HG3
 
 | | Human alpha-galactosidase catalytic mechanism 2. Substrate bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-galactosidase A, ... | | Authors: | Guce, A.I, Clark, N.E, Garman, S.C. | | Deposit date: | 2009-05-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic mechanism of human alpha-galactosidase.
J.Biol.Chem., 285, 2010
|
|
4L4V
 
 | | Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
3ES9
 
 | | NADPH-Cytochrome P450 Reductase in an Open Conformation | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hamdane, D, Xia, C, Im, S.-C, Zhang, H, Kim, J.-J, Waskell, L. | | Deposit date: | 2008-10-05 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and function of an NADPH-cytochrome P450 oxidoreductase in an open conformation capable of reducing cytochrome P450
J.Biol.Chem., 284, 2009
|
|
5X5U
 
 | |
2RFY
 
 | | Crystal structure of cellobiohydrolase from Melanocarpus albomyces complexed with cellobiose | | Descriptor: | Cellulose 1,4-beta-cellobiosidase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Parkkinen, T, Koivula, A, Vehmaanper, J, Rouvinen, J. | | Deposit date: | 2007-10-02 | | Release date: | 2008-09-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Melanocarpus albomyces cellobiohydrolase Cel7B in complex with cello-oligomers show high flexibility in the substrate binding
Protein Sci., 17, 2008
|
|
3EGJ
 
 | | N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae. | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase, NICKEL (II) ION, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of N-acetylglucosamine-6-phosphate deacetylase from Vibrio cholerae.
To be Published
|
|
7Q04
 
 | | Crystal structure of TPADO in a substrate-free state | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6IJC
 
 | | Structure of MMPA-CoA dehydrogenase from Roseovarius nubinhibens ISM | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-CoA dehydrogenase family protein | | Authors: | Shao, X, Yuan, Z.L, Cao, H.Y, Wang, P, Li, C.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
2IJ4
 
 | | Structure of the A264K mutant of cytochrome P450 BM3 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 | | Authors: | Toogood, H.S, Leys, D. | | Deposit date: | 2006-09-29 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and spectroscopic characterization of P450 BM3 mutants with unprecedented P450 heme iron ligand sets. New heme ligation states influence conformational equilibria in P450 BM3.
J.Biol.Chem., 282, 2007
|
|
