1QPW
 
 | | CRYSTAL STRUCTURE DETERMINATION OF PORCINE HEMOGLOBIN AT 1.8A RESOLUTION | | Descriptor: | OXYGEN MOLECULE, PORCINE HEMOGLOBIN (ALPHA SUBUNIT), PORCINE HEMOGLOBIN (BETA SUBUNIT), ... | | Authors: | Lu, T.-H, Panneerselvam, K, Liaw, Y.-C, Kan, P, Lee, C.-J. | | Deposit date: | 1999-05-30 | | Release date: | 1999-06-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of porcine haemoglobin.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1QQB
 
 | | PURINE REPRESSOR MUTANT-HYPOXANTHINE-PALINDROMIC OPERATOR COMPLEX | | Descriptor: | 5'-D(*TP*AP*CP*GP*CP*AP*AP*TP*CP*GP*AP*TP*TP*GP*CP*GP*T)-3', HYPOXANTHINE, PURINE NUCLEOTIDE SYNTHESIS REPRESSOR | | Authors: | Glasfeld, A, Koehler, A.N, Schumacher, M.A, Brennan, R.G. | | Deposit date: | 1999-06-01 | | Release date: | 1999-06-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions.
J.Mol.Biol., 291, 1999
|
|
1QR9
 
 | | INHIBITION OF HIV-1 INFECTIVITY BY THE GP41 CORE: ROLE OF A CONSERVED HYDROPHOBIC CAVITY IN MEMBRANE FUSION | | Descriptor: | GP41 ENVELOPE PROTEIN | | Authors: | Ji, H, Shu, W, Burling, F.T, Jiang, S.B, Lu, M. | | Deposit date: | 1999-06-18 | | Release date: | 1999-11-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion.
J.Virol., 73, 1999
|
|
3STK
 
 | |
3TF4
 
 | | ENDO/EXOCELLULASE:CELLOTRIOSE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN, beta-D-glucopyranose, ... | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
1QVN
 
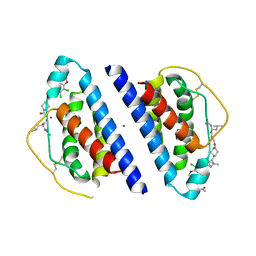 | | Structure of SP4160 Bound to IL-2 V69A | | Descriptor: | 2-GUANIDINO-4-METHYL-PENTANOIC ACID [2-(4-{5-[4-(4-ACETYLAMINO-BENZYLOXY)-2,3-DICHLORO-PHENYL]-2-METHYL-2H-PYRAZOL-3-YL}-PIPERIDIN-1-YL)-2-OXO-ETHYL]-AMIDE, Interleukin-2, ZINC ION | | Authors: | Thanos, C.D, Delano, W.L, Wells, J.A. | | Deposit date: | 2003-08-28 | | Release date: | 2005-04-05 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Hot-spot mimicry of a cytokine receptor by a small molecule.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
5ZC4
 
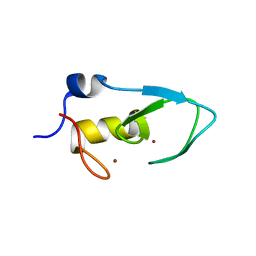 | |
1YQO
 
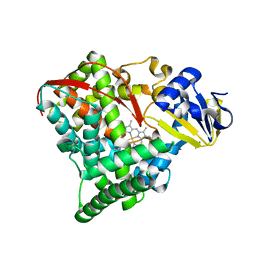 | | T268A mutant heme domain of flavocytochrome P450 BM3 | | Descriptor: | Bifunctional P-450:NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Clark, J.P, Miles, C.S, Mowat, C.G, Walkinshaw, M.D, Reid, G.A, Daff, S.N, Chapman, S.K. | | Deposit date: | 2005-02-02 | | Release date: | 2006-01-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of Thr268 and Phe393 in cytochrome P450 BM3.
J.Inorg.Biochem., 100, 2006
|
|
1TO5
 
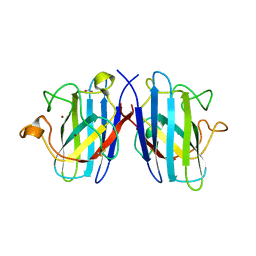 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | ACETATE ION, COPPER (II) ION, Superoxide dismutase, ... | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2KHS
 
 | | Solution structure of SNase121:SNase(111-143) complex | | Descriptor: | Nuclease, Thermonuclease | | Authors: | Geng, Y, Feng, Y, Xie, T, Shan, L, Wang, J. | | Deposit date: | 2009-04-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The native-like interactions between SNase121 and SNase(111-143) fragments induce the recovery of their native-like structures and the ability to degrade DNA.
Biochemistry, 48, 2009
|
|
8T1S
 
 | | Structure of the alpha-N-methyltransferase (SonM) and RiPP precursor (SonA with QSY deletion) heteromeric complex (bound to SAH) | | Descriptor: | Alpha-N-methyltransferase, Extradiol ring-cleavage dioxygenase LigAB LigA subunit domain-containing protein, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Crone, K.K, Jomori, T, Miller, F.S, Gralnick, J, Elias, M, Freeman, M.F. | | Deposit date: | 2023-06-03 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RiPP enzyme heterocomplex structure-guided discovery of a bacterial borosin alpha- N -methylated peptide natural product.
Rsc Chem Biol, 4, 2023
|
|
1LCE
 
 | | LACTOBACILLUS CASEI THYMIDYLATE SYNTHASE TERNARY COMPLEX WITH DUMP AND CH2THF | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5,10-METHYLENE-6-HYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Birdsall, D.L, Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-06-22 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refined structures of substrate-bound and phosphate-bound thymidylate synthase from Lactobacillus casei.
J.Mol.Biol., 232, 1993
|
|
3A6H
 
 | | W154A mutant creatininase | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3A6K
 
 | | The E122Q mutant creatininase, Mn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, MANGANESE (II) ION, ... | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3NYK
 
 | |
3A8U
 
 | |
2L9H
 
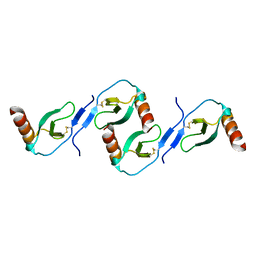 | | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data | | Descriptor: | C-C motif chemokine 5 | | Authors: | Wang, X, Watson, C.M, Sharp, J.S, Handel, T.M, Prestegard, J.H. | | Deposit date: | 2011-02-09 | | Release date: | 2011-06-22 | | Last modified: | 2011-08-24 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Oligomeric Structure of the Chemokine CCL5/RANTES from NMR, MS, and SAXS Data.
Structure, 19, 2011
|
|
2OX1
 
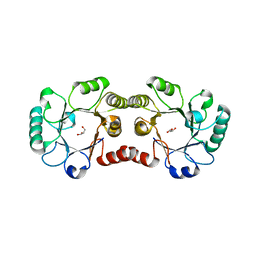 | | Archaeal Dehydroquinase | | Descriptor: | 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Gallagher, D.T, Smith, N.N. | | Deposit date: | 2007-02-19 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure and lability of archaeal dehydroquinase.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
1LQB
 
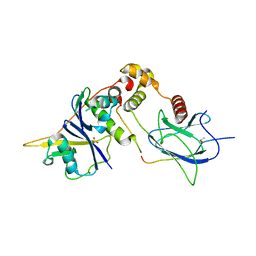 | | Crystal structure of a hydroxylated HIF-1 alpha peptide bound to the pVHL/elongin-C/elongin-B complex | | Descriptor: | Elongin B, Elongin C, Hypoxia-inducible factor 1 ALPHA, ... | | Authors: | Hon, W.C, Wilson, M.I, Harlos, K, Claridge, T.D, Schofield, C.J, Pugh, C.W, Maxwell, P.H, Ratcliffe, P.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2002-05-09 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL.
Nature, 417, 2002
|
|
1Z1R
 
 | | HIV-1 protease complexed with Macrocyclic peptidomimetic inhibitor 2 | | Descriptor: | 2-[(8S,11S)-11-{(1R)-1-HYDROXY-2-[ISOPENTYL(PHENYLSULFONYL)AMINO]ETHYL}-6,9-DIOXO-2-OXA-7,10-DIAZABICYCLO[11.2.2]HEPTADECA-1(15),13,16-TRIEN-8-YL]ACETAMIDE, Pol polyprotein, SULFATE ION | | Authors: | Martin, J.L, Begun, J, Schindeler, A, Wickramasinghe, W.A, Alewood, D, Alewood, P.F, Bergman, D.A, Brinkworth, R.I, Abbenante, G, March, D.R, Reid, R.C, Fairlie, D.P. | | Deposit date: | 2005-03-06 | | Release date: | 2005-03-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular recognition of macrocyclic peptidomimetic inhibitors by HIV-1 protease
Biochemistry, 38, 1999
|
|
3A6L
 
 | | E122Q mutant creatininase, Zn-Zn type | | Descriptor: | CHLORIDE ION, Creatinine amidohydrolase, ZINC ION | | Authors: | Nakajima, Y, Yamashita, K, Ito, K, Yoshimoto, T. | | Deposit date: | 2009-09-02 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase
J.Mol.Biol., 396, 2010
|
|
3ACC
 
 | | Crystal structure of hypoxanthine-guanine phosphoribosyltransferase with GMP from Thermus thermophilus HB8 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, GUANOSINE-5'-MONOPHOSPHATE, Hypoxanthine-guanine phosphoribosyltransferase | | Authors: | Kanagawa, M, Baba, S, Hirotsu, K, Kuramitsu, S, Yokoyama, S, Kawai, G, Sampei, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-12-30 | | Release date: | 2010-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structures of hypoxanthine-guanine phosphoribosyltransferase (TTHA0220) from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
4HNJ
 
 | |
2P9Q
 
 | | Crystal Structure of Phosphoglycerate Kinase-2 | | Descriptor: | Phosphoglycerate kinase, testis specific | | Authors: | Sawyer, G.M, Monzingo, A.F, Poteet, E.C, Robertus, J.D. | | Deposit date: | 2007-03-26 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray analysis of phosphoglycerate kinase 2, a sperm-specific isoform from Mus musculus.
Proteins, 71, 2007
|
|
1LCB
 
 | |
