5OB1
 
 | |
6SII
 
 | |
5O9F
 
 | | Crystal structure of R. ruber ADH-A, mutant Y294F, W295A, Y54F, F43S, H39Y | | Descriptor: | (2~{S})-2-methylpentanedioic acid, Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Relaxation of nonproductive binding and increased rate of coenzyme release in an alcohol dehydrogenase increases turnover with a nonpreferred alcohol enantiomer.
FEBS J., 284, 2017
|
|
6SIS
 
 | | Crystal structure of macrocyclic PROTAC 1 in complex with the second bromodomain of human Brd4 and pVHL:ElonginC:ElonginB | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Hughes, S.J, Testa, A, Ciulli, A. | | Deposit date: | 2019-08-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure-Based Design of a Macrocyclic PROTAC.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5OCM
 
 | | Imine Reductase from Streptosporangium roseum in complex with NADP+ and 2,2,2-trifluoroacetophenone hydrate | | Descriptor: | 2,2,2-trifluoromethyl acetophenone hydrate, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Sharma, M, Nestl, B, Grogan, G. | | Deposit date: | 2017-07-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | New imine-reducing enzymes from beta-hydroxyacid dehydrogenases by single amino acid substitutions.
Protein Eng. Des. Sel., 31, 2018
|
|
6SLG
 
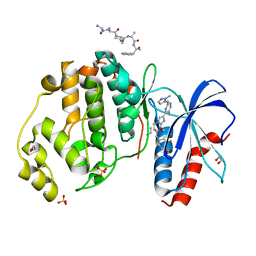 | | HUMAN ERK2 WITH ERK1/2 INHIBITOR, AZD0364. | | Descriptor: | (6~{R})-7-[[3,4-bis(fluoranyl)phenyl]methyl]-6-(methoxymethyl)-2-[5-methyl-2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-5,6-dihydroimidazo[1,2-a]pyrazin-8-one, 1,2-ETHANEDIOL, ERK-tide, ... | | Authors: | Breed, J, Phillips, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Discovery of a Potent and Selective Oral Inhibitor of ERK1/2 (AZD0364) That Is Efficacious in Both Monotherapy and Combination Therapy in Models of Nonsmall Cell Lung Cancer (NSCLC).
J.Med.Chem., 62, 2019
|
|
7CNX
 
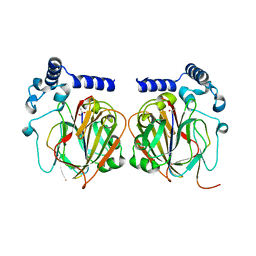 | | Crystal structure of Apo PSD from E. coli (2.63 A) | | Descriptor: | Phosphatidylserine decarboxylase alpha chain, Phosphatidylserine decarboxylase beta chain | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
5OE0
 
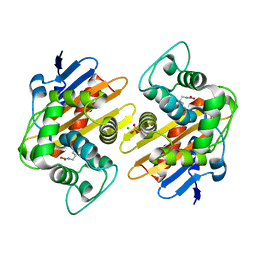 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-181 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Lund, B.A, Carlsen, T.J.O, Leiros, H.K.S, Thomassen, A.M. | | Deposit date: | 2017-07-07 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.0500083 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7CNY
 
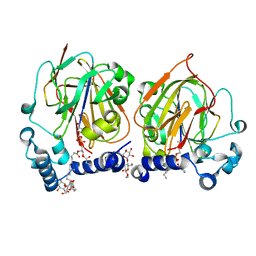 | | Crystal structure of 8PE bound PSD from E. coli (2.12 A) | | Descriptor: | 1,2-Dioctanoyl-SN-Glycero-3-Phosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
6SFG
 
 | | CRYSTAL STRUCTURE OF DHQ1 FROM SALMONELLA TYPHI COVALENTLY MODIFIED BY COMPOUND 9 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, (1~{S},3~{R},4~{S},5~{R})-3-methyl-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic Acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
5OE6
 
 | |
7CNZ
 
 | | Crystal structure of 10PE bound PSD from E. coli (2.70 A) | | Descriptor: | 1,2-DIDECANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, PHOSPHATE ION, Phosphatidylserine decarboxylase alpha chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
6SMF
 
 | |
5OF6
 
 | | Cu nitrite reductase serial data at varying temperatures 190K 0.48MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-10 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
7CNW
 
 | | Crystal structure of Apo PSD from E. coli (1.90 A) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Phosphatidylserine decarboxylase alpha chain, Phosphatidylserine decarboxylase beta chain, ... | | Authors: | Kim, J, Cho, G. | | Deposit date: | 2020-08-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into phosphatidylethanolamine formation in bacterial membrane biogenesis.
Sci Rep, 11, 2021
|
|
5OFG
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.06MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
7CNB
 
 | |
5OHE
 
 | | Globin sensor domain of AfGcHK (FeIII form) in complex with cyanide | | Descriptor: | CHLORIDE ION, CYANIDE ION, Globin-coupled histidine kinase, ... | | Authors: | Skalova, T, Kolenko, P, Dohnalek, J, Stranava, M, Martinkova, M. | | Deposit date: | 2017-07-16 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Coordination and redox state-dependent structural changes of the heme-based oxygen sensor AfGcHK associated with intraprotein signal transduction.
J. Biol. Chem., 292, 2017
|
|
5O5X
 
 | | Crystal structure of Thermococcus litoralis ADP-dependent glucokinase (GK) | | Descriptor: | ADP-dependent glucokinase,ADP-dependent glucokinase,ADP-dependent glucokinase, GLYCEROL, SULFATE ION | | Authors: | Herrera-Morande, A, Castro-Fernandez, V, Merino, F, Ramirez-Sarmiento, C.A, Fernandez, F.J, Guixe, V, Vega, M.C. | | Deposit date: | 2017-06-02 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Protein topology determines substrate-binding mechanism in homologous enzymes.
Biochim Biophys Acta Gen Subj, 1862, 2018
|
|
7CKG
 
 | | Crystal structure of TMSiPheRS complexed with TMSiPhe | | Descriptor: | 4-(trimethylsilyl)-L-phenylalanine, Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
6SKS
 
 | |
5OJJ
 
 | | Crystal structure of the Zn-bound ubiquitin-conjugating enzyme Ube2T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Ubiquitin-conjugating enzyme E2 T, ... | | Authors: | Morreale, F.E, Testa, A, Chaugule, V.K, Bortoluzzi, A, Ciulli, A, Walden, H. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mind the Metal: A Fragment Library-Derived Zinc Impurity Binds the E2 Ubiquitin-Conjugating Enzyme Ube2T and Induces Structural Rearrangements.
J. Med. Chem., 60, 2017
|
|
7CKH
 
 | | Crystal structure of TMSiPheRS | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79492676 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
5O6M
 
 | |
7CE2
 
 | | The Crystal structure of TeNT Hc complexed with neutralizing antibody | | Descriptor: | Tetanus toxin, neutralizing antibody heavy chain, neutralizing antibody light chain | | Authors: | Wang, X, Wang, Y, Wu, C, Yu, J, Liao, H. | | Deposit date: | 2020-06-21 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis of tetanus toxin neutralization by native human monoclonal antibodies.
Cell Rep, 35, 2021
|
|
