7F07
 
 | | Autonomous VH domain that interacts with eIF4E at the Capped mRNA Binding site. | | Descriptor: | Eukaryotic translation initiation factor 4E, VH domain (VH-DiFCAP-01) | | Authors: | Brown, C.J, Frosi, Y, Ng, S, Lin, Y.C. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
6D6R
 
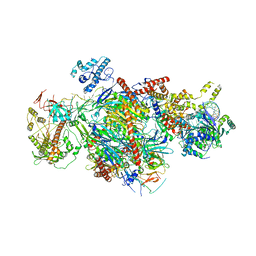 | |
6D6Q
 
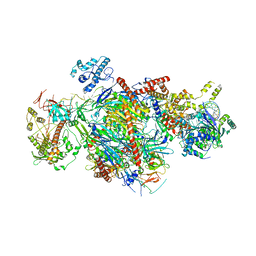 | |
8YUQ
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and dA4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUP
 
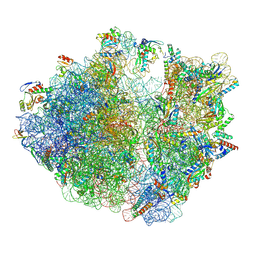 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and A4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-28 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUR
 
 | | E. coli 70S ribosome complexed with P. putida tRNAIle2 and Am4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
8YUS
 
 | | E. coli 70S ribosome complexed with P.putida tRNAIle2 and A(F)4 mRNA | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Akiyama, N, Ishiguro, K, Shirouzu, M, Suzuki, T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-11-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | A tRNA modification with aminovaleramide facilitates AUA decoding in protein synthesis.
Nat.Chem.Biol., 21, 2025
|
|
6ZTJ
 
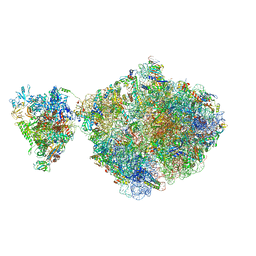 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (38 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTN
 
 | | E. coli 70S-RNAP expressome complex in NusG-coupled state (42 nt intervening mRNA) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTP
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 6 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
8W8F
 
 | | human co-transcriptional RNA capping enzyme RNGTT-CMTR1 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (36-MER), DNA (45-MER), ... | | Authors: | Li, Y, Wang, Q, Xu, Y, Li, Z. | | Deposit date: | 2023-09-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures of co-transcriptional RNA capping enzymes on paused transcription complex.
Nat Commun, 15, 2024
|
|
6ZTL
 
 | | E. coli 70S-RNAP expressome complex in collided state bound to NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTO
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 1 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-23 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZU1
 
 | | E. coli 70S-RNAP expressome complex in uncoupled state 2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-21 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
6ZTM
 
 | | E. coli 70S-RNAP expressome complex in collided state without NusG | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Webster, M.W, Takacs, M, Weixlbaumer, A. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of transcription-translation coupling and collision in bacteria.
Science, 369, 2020
|
|
3J81
 
 | | CryoEM structure of a partial yeast 48S preinitiation complex | | Descriptor: | 18S rRNA, MAGNESIUM ION, METHIONINE, ... | | Authors: | Hussain, T, Llacer, J.L, Fernandez, I.S, Savva, C.G, Ramakrishnan, V. | | Deposit date: | 2014-08-29 | | Release date: | 2014-11-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural changes enable start codon recognition by the eukaryotic translation initiation complex.
Cell(Cambridge,Mass.), 159, 2014
|
|
6ZSE
 
 | | Human mitochondrial ribosome in complex with mRNA, A/P-tRNA and P/E-tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
7AC7
 
 | | Structure of accomodated trans-translation complex on E. Coli stalled ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | Deposit date: | 2020-09-10 | | Release date: | 2021-08-18 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
8RAN
 
 | |
4KZZ
 
 | |
6RFL
 
 | | Structure of the complete Vaccinia DNA-dependent RNA polymerase complex | | Descriptor: | DNA-dependent RNA polymerase subunit rpo132, DNA-dependent RNA polymerase subunit rpo147, DNA-dependent RNA polymerase subunit rpo18, ... | | Authors: | Grimm, C, Hillen, S.H, Bedenk, K, Bartuli, J, Neyer, S, Zhang, Q, Huettenhofer, A, Erlacher, M, Dienemann, C, Schlosser, A, Urlaub, H, Boettcher, B, Szalay, A.A, Cramer, P, Fischer, U. | | Deposit date: | 2019-04-15 | | Release date: | 2019-12-11 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural Basis of Poxvirus Transcription: Vaccinia RNA Polymerase Complexes.
Cell, 179, 2019
|
|
8OVE
 
 | |
5VSU
 
 | | Structure of yeast U6 snRNP with 2'-phosphate terminated U6 RNA | | Descriptor: | Saccharomyces cerevisiae strain T8 chromosome XII sequence, U4/U6 snRNA-associated-splicing factor PRP24, U6 snRNA-associated Sm-like protein LSm2, ... | | Authors: | Montemayor, E.J. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Architecture of the U6 snRNP reveals specific recognition of 3'-end processed U6 snRNA.
Nat Commun, 9, 2018
|
|
5NPM
 
 | | CRYSTAL STRUCTURE OF MUTANT RIBOSOMAL PROTEIN TTHL1 LACKING 8 N-TERMINAL RESIDUES IN COMPLEX WITH 80NT 23S RNA FROM THERMUS THERMOPHILUS | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L1, MALONATE ION, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, T.V, Nevskaya, N.A, Nikonov, S.V, Garber, M.B. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | [Influence of Nonconserved Regions of L1 Protuberance of Thermus thermophilus Ribosome on the Affinity of L1 Protein to 23s rRNA].
Mol.Biol.(Moscow), 52, 2018
|
|
8WIC
 
 | | Cryo- EM structure of Mycobacterium smegmatis 50S ribosomal subunit (body 1) of 70S ribosome, E- tRNA and RafH. | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Kumar, N, Sharma, S, Kaushal, P.S. | | Deposit date: | 2023-09-24 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo- EM structure of the mycobacterial 70S ribosome in complex with ribosome hibernation promotion factor RafH.
Nat Commun, 15, 2024
|
|
