2VRN
 
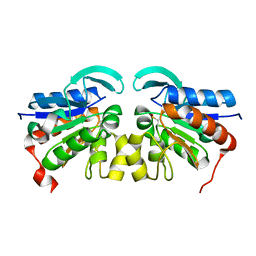 | | The structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily | | Descriptor: | MAGNESIUM ION, PROTEASE I | | Authors: | Fioravanti, E, Dura, M.A, Lascoux, D, Micossi, E, McSweeney, S. | | Deposit date: | 2008-04-09 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily.
Biochemistry, 47, 2008
|
|
9IKI
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitrous Oxide-bound Fully Reduced State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Ide, T, Shinzawa-Itoh, K. | | Deposit date: | 2024-06-27 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
5L5O
 
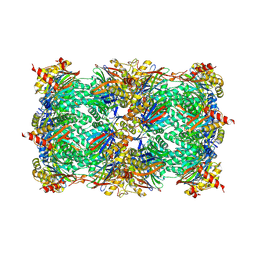 | | Yeast 20S proteasome with human beta5i (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
7FAI
 
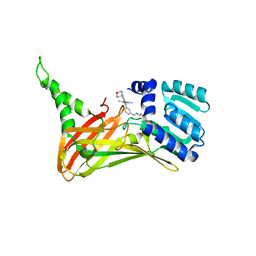 | | CARM1 bound with compound 9 | | Descriptor: | Histone-arginine methyltransferase CARM1, N'-[[3-[4-(3,5-dimethyl-1,2-oxazol-4-yl)-5-methyl-6-(oxan-4-ylamino)pyrimidin-2-yl]phenyl]methyl]-N-methyl-ethane-1,2-diamine | | Authors: | Cao, D.Y, Li, J, Xiong, B. | | Deposit date: | 2021-07-06 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09749269 Å) | | Cite: | Structure-Based Discovery of Potent CARM1 Inhibitors for Solid Tumor and Cancer Immunology Therapy.
J.Med.Chem., 64, 2021
|
|
5L65
 
 | |
6U7K
 
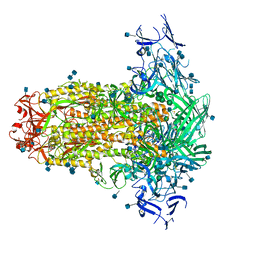 | | Prefusion structure of PEDV spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wrapp, D, McLellan, J.S. | | Deposit date: | 2019-09-03 | | Release date: | 2019-09-18 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation.
J.Virol., 93, 2019
|
|
3I8W
 
 | |
5L5H
 
 | |
5NBP
 
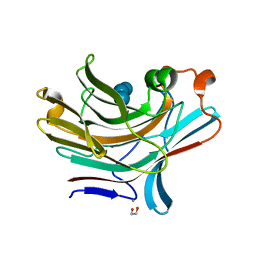 | | Bacteroides ovatus mixed linkage glucan PUL (MLGUL) GH16 in complex with G4G4G3G Product | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glycosyl hydrolase family 16, ... | | Authors: | Hemsworth, G.R, Tamura, K, Dejean, G, Rogers, T.E, Pudlo, N.A, Urs, K, Jain, N, Martens, E.C, Brumer, H, Davies, G.J. | | Deposit date: | 2017-03-02 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Mechanism by which Prominent Human Gut Bacteroidetes Utilize Mixed-Linkage Beta-Glucans, Major Health-Promoting Cereal Polysaccharides.
Cell Rep, 21, 2017
|
|
9IKG
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Dioxide-bound Fully Reduced State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2024-06-27 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
9IKH
 
 | | Bovine Heart Cytochrome c Oxidase in the Nitrous Oxide-bound Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Ide, T, Shinzawa-Itoh, K. | | Deposit date: | 2024-06-27 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
9IKF
 
 | | Bovine Heart Cytochrome c Oxidase in the Carbon Dioxide-bound Fully Oxidized State | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, 1,2-ETHANEDIOL, ... | | Authors: | Muramoto, K, Shinzawa-Itoh, K. | | Deposit date: | 2024-06-27 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding sites of carbon dioxide, nitrous oxide, and xenon reveal a putative exhaust channel for bovine cytochrome c oxidase.
J.Biol.Chem., 301, 2025
|
|
5UE8
 
 | | The crystal structure of Munc13-1 C1C2BMUN domain | | Descriptor: | CHLORIDE ION, Protein unc-13 homolog A, ZINC ION | | Authors: | Tomchick, D.R, Rizo, J, Xu, J. | | Deposit date: | 2016-12-29 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN.
Elife, 6, 2017
|
|
1L4F
 
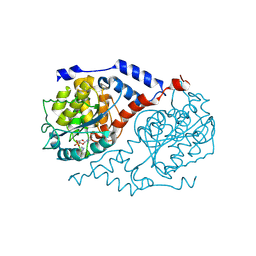 | | The crystal structure of CobT complexed with 4,5-dimethyl-1,2-phenylenediamine and nicotinate mononucleotide | | Descriptor: | 4,5-DIMETHYL-1,2-PHENYLENEDIAMINE, NICOTINATE MONONUCLEOTIDE, Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-06 | | Release date: | 2002-09-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
4F4E
 
 | |
1QD6
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI | | Descriptor: | 1-HEXADECANOSULFONIC ACID, CALCIUM ION, OUTER MEMBRANE PHOSPHOLIPASE (OMPLA), ... | | Authors: | Snijder, H.J, Ubarretxena-Belandia, I, Blaauw, M, Kalk, K.H, Verheij, H.M, Egmond, M.R, Dekker, N, Dijkstra, B.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for dimerization-regulated activation of an integral membrane phospholipase.
Nature, 401, 1999
|
|
4BXH
 
 | | Resolving the activation site of positive regulators in plant phosphoenolpyruvate carboxylase | | Descriptor: | 1,2-ETHANEDIOL, C4 PHOSPHOENOLPYRUVATE CARBOXYLASE, SULFATE ION | | Authors: | Schlieper, D, Foerster, K, Paulus, J.K, Groth, G. | | Deposit date: | 2013-07-11 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Resolving the Activation Site of Positive Regulators in Plant Phosphoenolpyruvate Carboxylase.
Mol.Plant, 7, 2014
|
|
7EV2
 
 | |
4OY2
 
 | | Crystal structure of TAF1-TAF7, a TFIID subcomplex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 7, ... | | Authors: | Bhattacharya, S, Lou, X, Rajashankar, K, Jacobson, R.H, Webb, P. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional insight into TAF1-TAF7, a subcomplex of transcription factor II D.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3A21
 
 | | Crystal Structure of Streptomyces avermitilis beta-L-Arabinopyranosidase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Fujimoto, Z, Ichinose, H, Kaneko, S. | | Deposit date: | 2009-04-27 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.
J.Biol.Chem., 284, 2009
|
|
7C67
 
 | | Crystal structure of beta-glycosides-binding protein of ABC transporter in a closed state bound to cellotriose | | Descriptor: | 1,2-ETHANEDIOL, CARBONATE ION, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
7F1D
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(4R,5R,6R)-2-amino-5-fluoro-4,6-dimethyl-5,6-dihydro-4H-1,3-thiazin-4-yl]-4-fluorophenyl}-2H,3H-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Descriptor: | Beta-secretase 1, IODIDE ION, N-[3-[(4R,5R,6R)-2-azanyl-5-fluoranyl-4,6-dimethyl-5,6-dihydro-1,3-thiazin-4-yl]-4-fluoranyl-phenyl]-2,3-dihydro-[1,4]dioxino[2,3-c]pyridine-7-carboxamide | | Authors: | Ueno, T, Matsuoka, E, Asada, N, Yamamoto, S, Kanegawa, N, Ito, M, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2021-06-09 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of Extremely Selective Fused Pyridine-Derived beta-Site Amyloid Precursor Protein-Cleaving Enzyme (BACE1) Inhibitors with High In Vivo Efficacy through 10s Loop Interactions.
J.Med.Chem., 64, 2021
|
|
4FDU
 
 | | Crystal Structure of a Multiple Inositol Polyphosphate Phosphatase | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-HEXASULPHATE, Putative multiple inositol polyphosphate histidine phosphatase 1 | | Authors: | Li, A.W.H, Hemmings, A.M. | | Deposit date: | 2012-05-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | A Bacterial Homolog of a Eukaryotic Inositol Phosphate Signaling Enzyme Mediates Cross-kingdom Dialog in the Mammalian Gut.
Cell Rep, 6, 2014
|
|
1L8P
 
 | | Mg-phosphonoacetohydroxamate complex of S39A yeast enolase 1 | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID, enolase 1 | | Authors: | Poyner, R.R, Larsen, T.M, Wong, S.W, Reed, G.H. | | Deposit date: | 2002-03-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase.
Arch.Biochem.Biophys., 401, 2002
|
|
3IKF
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with FOL fragment 717, imidazo[2,,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
