4BG4
 
 | | Crystal structure of Litopenaeus vannamei arginine kinase in a ternary analog complex with arginine, ADP-Mg and NO3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, ARGININE KINASE, ... | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, GBrieba, L, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-03-22 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
4W61
 
 | |
4AYS
 
 | | The Structure of Amylosucrase from D. radiodurans | | Descriptor: | AMYLOSUCRASE | | Authors: | Skov, L.K, Pizzut, S, Remaud-Simeon, M, Gajhede, M, Mirza, O. | | Deposit date: | 2012-06-21 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | The Structure of Amylosucrase from Deinococcus Radiodurans Has an Unusual Open Active-Site Topology
Acta Crystallogr.,Sect.F, 69, 2013
|
|
8B3C
 
 | | Chalcone synthase from Hordeum vulgare complexed with CoA and eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, Chalcone synthase 2 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Plant Polyketide Synthase for the Biosynthesis of Methylated Flavonoids.
J.Agric.Food Chem., 72, 2024
|
|
4V82
 
 | | Crystal structure of cyanobacterial Photosystem II in complex with terbutryn | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Gabdulkhakov, A, Broser, M, Guskov, A, Kern, J, Glockner, C, Muh, F, Saenger, W, Zouni, A. | | Deposit date: | 2010-11-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of cyanobacterial photosystem II Inhibition by the herbicide terbutryn
J.Biol.Chem., 286, 2011
|
|
2Z0Z
 
 | | Crystal structure of putative acetyltransferase | | Descriptor: | Putative uncharacterized protein TTHA1799, SULFATE ION | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Genetic Encoding of 3-Iodo-l-Tyrosine in Escherichia coli for Single-Wavelength Anomalous Dispersion Phasing in Protein Crystallography
Structure, 17, 2009
|
|
4BHL
 
 | | Crystal structure of Litopenaeus vannamei arginine kinase in binary complex with arginine | | Descriptor: | ARGININE, ARGININE KINASE, BETA-MERCAPTOETHANOL | | Authors: | Lopez-Zavala, A.A, Garcia-Orozco, K.D, Carrasco-Miranda, J.S, Hernandez-Flores, J.M, Sugich-Miranda, R, Velazquez-Contreras, E.F, Criscitiello, M.F, Brieba, L.G, Rudino-Pinera, E, Sotelo-Mundo, R.R. | | Deposit date: | 2013-04-03 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Shrimp Arginine Kinase in Binary Complex with Arginine-A Molecular View of the Phosphagen Precursor Binding to the Enzyme.
J.Bioenerg.Biomembr., 45, 2013
|
|
6I3D
 
 | |
7Z07
 
 | |
7Z08
 
 | |
7Z0B
 
 | |
2Z11
 
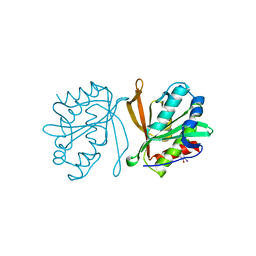 | | Crystal structure of putative acetyltransferase | | Descriptor: | Ribosomal-protein-alanine acetyltransferase | | Authors: | Murayama, K, Kato-Murayama, M, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2007-11-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative acetyltransferase
To be Published
|
|
2J9F
 
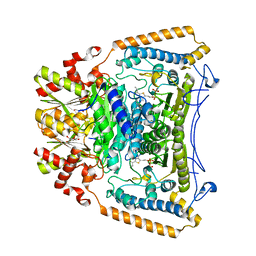 | | Human branched-chain alpha-ketoacid dehydrogenase-decarboxylase E1b | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, C2-1-HYDROXY-3-METHYL-PROPYL-THIAMIN DIPHOSPHATE, ... | | Authors: | Jun, L, Machius, M, Chuang, J.L, Wynn, R.M, Chuang, D.T. | | Deposit date: | 2006-11-07 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The Two Active Sites in Human Branched-Chain Alpha- Keto Acid Dehydrogenase Operate Independently without an Obligatory Alternating-Site Mechanism.
J.Biol.Chem., 282, 2007
|
|
2IEA
 
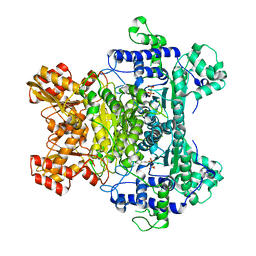 | | E. coli pyruvate dehydrogenase | | Descriptor: | MAGNESIUM ION, Pyruvate dehydrogenase E1 component, THIAMINE DIPHOSPHATE | | Authors: | Furey, W, Arjunan, P. | | Deposit date: | 2006-09-18 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the pyruvate dehydrogenase multienzyme complex E1 component from Escherichia coli at 1.85 A resolution.
Biochemistry, 41, 2002
|
|
7YTL
 
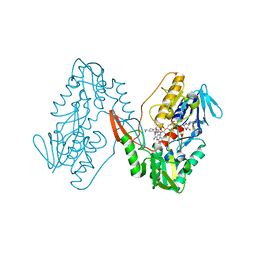 | | Structure of a oxidoreductase in complex with quinone | | Descriptor: | Apoptosis inducing factor mitochondria associated 2, FLAVIN-ADENINE DINUCLEOTIDE, UBIQUINONE-1 | | Authors: | Lv, Y, Sun, Q, Wang, Q, Zhu, D. | | Deposit date: | 2022-08-15 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural insights into FSP1 catalysis and ferroptosis inhibition.
Nat Commun, 14, 2023
|
|
7ZYV
 
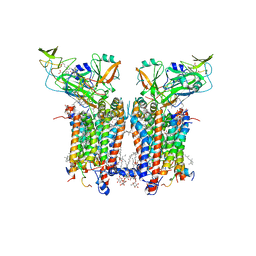 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.13 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, ... | | Authors: | Sarewicz, M, Szwalec, M, Pintscher, S, Indyka, P, Rawski, M, Pietras, R, Mielecki, B, Koziej, L, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-05-25 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
6SGN
 
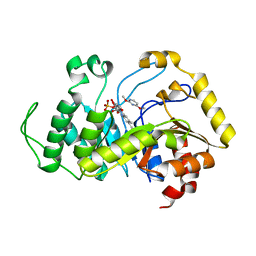 | | Crystal structure of monooxygenase RutA complexed with 2,4-dimethoxypyrimidine. | | Descriptor: | 2,4-dimethoxypyrimidine, FLAVIN MONONUCLEOTIDE, Pyrimidine monooxygenase RutA | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6SGG
 
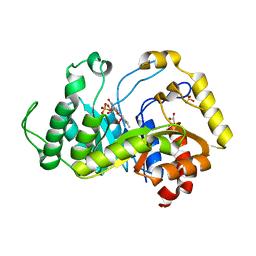 | | Crystal structure of monooxygenase RutA complexed with dioxygen under 1.5 MPa / 15 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-04 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6SGL
 
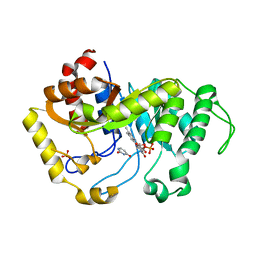 | | Crystal structure of monooxygenase RutA complexed with Uracil under atmospheric pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, Pyrimidine monooxygenase RutA, SULFATE ION, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-08-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6SY1
 
 | | Crystal structure of the human 2-oxoadipate dehydrogenase DHTKD1 (E1) | | Descriptor: | MAGNESIUM ION, Probable 2-oxoglutarate dehydrogenase E1 component DHKTD1, mitochondrial, ... | | Authors: | Bezerra, G.A, Foster, W, Shrestha, L, Pena, I.A, Coker, J, Kolker, S, Nicola, B.B, von Delft, F, Edwards, A, Arrowsmith, C, Bountra, C, Yue, W.W, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure and interaction studies of human DHTKD1 provide insight into a mitochondrial megacomplex in lysine catabolism.
Iucrj, 7, 2020
|
|
6SGM
 
 | |
6VEF
 
 | |
6TEG
 
 | | Crystal structure of monooxygenase RutA complexed with uracil and dioxygen under 1.5 MPa / 15 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, OXYGEN MOLECULE, Pyrimidine monooxygenase RutA, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-11-11 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6TEF
 
 | | Crystal structure of monooxygenase RutA complexed with dioxygen under 0.5 MPa / 5 bars of oxygen pressure. | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, OXYGEN MOLECULE, ... | | Authors: | Saleem-Batcha, R, Matthews, A, Teufel, R. | | Deposit date: | 2019-11-11 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Aminoperoxide adducts expand the catalytic repertoire of flavin monooxygenases.
Nat.Chem.Biol., 16, 2020
|
|
6TLK
 
 | | Structure of methylene-tetrahydromethanopterin dehydrogenase from Methylorubrum extorquens AM1 in an open conformation containing NADP+ and methylene-H4MPT | | Descriptor: | 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, Bifunctional protein MdtA, CHLORIDE ION, ... | | Authors: | Wagner, T, Huang, G, Demmer, U, Warkentin, E, Ermler, U, Shima, S. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Hydride Transfer Process in NADP-dependent Methylene-tetrahydromethanopterin Dehydrogenase.
J.Mol.Biol., 432, 2020
|
|
