5O06
 
 | | Crystal structure of APO form of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus | | Descriptor: | 1,2-ETHANEDIOL, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
7COM
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Boceprevir (space group P212121) | | Descriptor: | 3C-like proteinase, boceprevir (bound form) | | Authors: | Zeng, R, Qiao, J.X, Wang, Y.F, Li, Y.S, Yao, R, Liu, J.M, Zhou, Y.L, Chen, P, Yang, S.Y, Lei, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro inhibitors with antiviral activity in a transgenic mouse model.
Science, 371, 2021
|
|
5O3H
 
 | | Human Brd2(BD2) mutant in complex with 9-ME-Am1 | | Descriptor: | (2~{R})-2-[(4~{S})-6-(4-chlorophenyl)-9-methoxy-1-methyl-4~{H}-[1,2,4]triazolo[4,3-a][1,4]benzodiazepin-4-yl]-~{N}-ethyl-propanamide, (2~{S})-1-[(2~{S})-2-oxidanylpropoxy]propan-2-ol, Bromodomain-containing protein 2, ... | | Authors: | Chan, K.-H, Runcie, A.C, Ciulli, A. | | Deposit date: | 2017-05-23 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Optimization of a "bump-and-hole" approach to allele-selective BET bromodomain inhibition.
Chem Sci, 9, 2018
|
|
7C7G
 
 | |
5O0B
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-Bromo-1H-indazole-5-carboxylic acid (Fragment 2) | | Descriptor: | 3-bromanyl-1~{H}-indazole-5-carboxylic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.735 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
6S4P
 
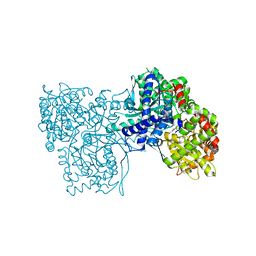 | | The crystal structure of glycogen phosphorylase in complex with 13 | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-(4-naphthalen-2-yl-1,3-thiazol-2-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-28 | | Release date: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6S58
 
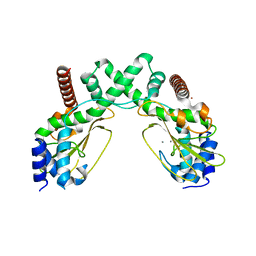 | | AvaII restriction endonuclease in the absence of nucleic acids | | Descriptor: | CALCIUM ION, Type II site-specific deoxyribonuclease, UNKNOWN ATOM OR ION | | Authors: | Kisiala, M, Kowalska, M, Korza, H, Czapinska, H, Bochtler, M. | | Deposit date: | 2019-06-30 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Restriction endonucleases that cleave RNA/DNA heteroduplexes bind dsDNA in A-like conformation.
Nucleic Acids Res., 48, 2020
|
|
7C83
 
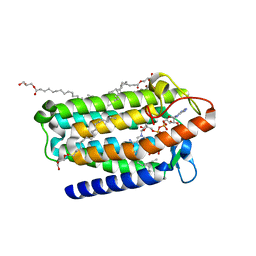 | | Crystal structure of an integral membrane steroid 5-alpha-reductase PbSRD5A | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-oxo-5-alpha-steroid 4-dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ren, R.B, Han, Y.F, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-05-28 | | Release date: | 2021-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Nat Commun, 12, 2021
|
|
6S5H
 
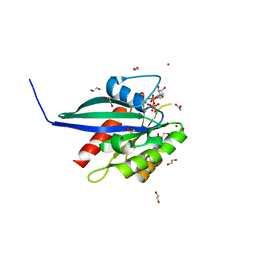 | | Structure of the human RAB38 in complex with GTP | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Diaz-Saez, L, Jung, S, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the human RAB38 in complex with GTP
To Be Published
|
|
6TZS
 
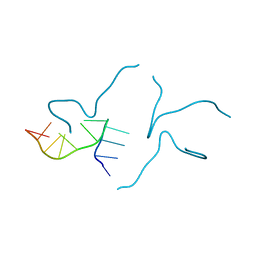 | | A DNA i-motif/duplex hybrid | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*CP*TP*GP*(CBR)P*AP*A)-3') | | Authors: | Chu, B, Paukstelis, P.J. | | Deposit date: | 2019-08-13 | | Release date: | 2019-10-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A DNA G-quadruplex/i-motif hybrid.
Nucleic Acids Res., 47, 2019
|
|
7CT3
 
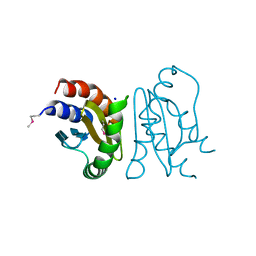 | | Crystal Structure of MglC from Myxococcus xanthus | | Descriptor: | Mutual gliding motility protein C (MglC), SODIUM ION | | Authors: | Thakur, K.G, Kapoor, S, Kodesia, A. | | Deposit date: | 2020-08-17 | | Release date: | 2021-01-27 | | Last modified: | 2021-07-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
J.Biol.Chem., 296, 2021
|
|
6TUU
 
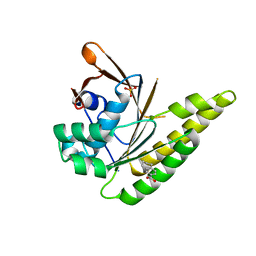 | | Leishmania infantum Rad51 surrogate LiRadA10 in complex with 5,6,7,8-tetrahydro-2-naphthoic acid | | Descriptor: | 5,6,7,8-tetrahydronaphthalene-2-carboxylic acid, CHLORIDE ION, DNA repair and recombination protein RadA, ... | | Authors: | Pantelejevs, T, Hyvonen, M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of dedicated crystallographic systems for structure-guided drug discovery
To Be Published
|
|
5O5A
 
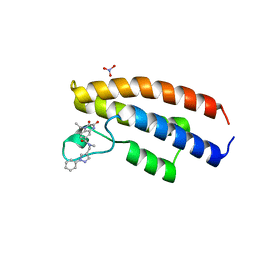 | | Crystal structure of the human BRPF1 bromodomain in complex with BZ032 | | Descriptor: | 1-ethyl-~{N}-methyl-2,3-bis(oxidanylidene)-~{N}-[(1-phenylpyrazol-4-yl)methyl]-4~{H}-quinoxaline-6-carboxamide, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-06-01 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based discovery of selective BRPF1 bromodomain inhibitors.
Eur J Med Chem, 155, 2018
|
|
5O2T
 
 | | Human KRAS in complex with darpin K27 | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Debreczeni, J.E, Guillard, S, Kolasinska-Zwierz, P, Breed, J, Zhang, J, Bery, N, Marwood, R, Tart, J, Stocki, P, Mistry, B, Phillips, C, Rabbitts, T, Jackson, R, Minter, R. | | Deposit date: | 2017-05-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and functional characterization of a DARPin which inhibits Ras nucleotide exchange.
Nat Commun, 8, 2017
|
|
5O30
 
 | |
7BR4
 
 | |
5O5U
 
 | | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027 | | Descriptor: | (2~{S})-2-[[(2~{S})-6-[[(2~{S})-2-[[4-[[[(2~{S})-2-[[(2~{R})-2-[(6-fluoranylpyridin-3-yl)carbonylamino]-3-sulfo-propanoyl]amino]-3-sulfo-propanoyl]amino]methyl]phenyl]carbonylamino]-3-naphthalen-2-yl-propanoyl]amino]-1-oxidanyl-1-oxidanylidene-hexan-2-yl]carbamoylamino]pentanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Novakova, Z, Barinka, C, Motlova, L. | | Deposit date: | 2017-06-02 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-ray structure of human glutamate carboxypeptidase II (GCPII) in complex with a urea based inhibitor PSMA 1027
To Be Published
|
|
6U1K
 
 | | Thermus thermophilus D-alanine-D-alanine ligase in complex with ADP, carbonate, D-alanine-D-alanine, Mg2+ and K+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CARBONATE ION, D-ALANINE, ... | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2019-08-15 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | d-Alanine-d-alanine ligase as a model for the activation of ATP-grasp enzymes by monovalent cations.
J.Biol.Chem., 295, 2020
|
|
6TVR
 
 | | Crystal structure of the haemagglutinin mutant (Gln226Leu) from an H10N7 seal influenza virus isolated in Germany | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
6U28
 
 | | Crystal structure of 1918 NS1-ED W187A in complex with the p85-beta-iSH2 domain of human PI3K | | Descriptor: | Non-structural protein 1, Phosphatidylinositol 3-kinase regulatory subunit beta | | Authors: | Cho, J.H, Zhao, B, Savage, N, Li, P. | | Deposit date: | 2019-08-19 | | Release date: | 2020-04-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of a host protein by NS1 of pandemic and seasonal influenza A viruses.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BJ6
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | (3~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-3-(4-chlorophenyl)-4-fluoranyl-3-[[1-(hydroxymethyl)cyclopropyl]methoxy]-6-(2-oxidanylpropan-2-yl)isoindol-1-one, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
5O7D
 
 | | The X-ray structure of human R38M phosphoglycerate kinase 1 mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphoglycerate kinase 1 | | Authors: | Ilari, A, Fiorillo, A, Cipollone, A, Petrosino, M. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The phosphoglycerate kinase 1 variants found in carcinoma cells display different catalytic activity and conformational stability compared to the native enzyme.
PLoS ONE, 13, 2018
|
|
5O7I
 
 | | ERK5 in complex with a pyrrole inhibitor | | Descriptor: | 4-(2-bromanyl-6-fluoranyl-phenyl)carbonyl-~{N}-pyridin-3-yl-1~{H}-pyrrole-2-carboxamide, DIMETHYL SULFOXIDE, Mitogen-activated protein kinase 7 | | Authors: | Tucker, J.A, Heptinstall, A, Myers, S. | | Deposit date: | 2017-06-08 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Identification of a novel orally bioavailable ERK5 inhibitor with selectivity over p38 alpha and BRD4.
Eur.J.Med.Chem., 178, 2019
|
|
5O7P
 
 | | HER3 in complex with Fab MF3178 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MF3178 FAB heavy chain, MF3178 FAB light chain, ... | | Authors: | De Nardis, C, Gros, P. | | Deposit date: | 2017-06-09 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Unbiased Combinatorial Screening Identifies a Bispecific IgG1 that Potently Inhibits HER3 Signaling via HER2-Guided Ligand Blockade.
Cancer Cell, 33, 2018
|
|
7BU1
 
 | |
