6SAK
 
 | |
5NNN
 
 | |
5NQ0
 
 | | Porcine (Sus scrofa) Major Histocompatibility Complex, class I, presenting DFEREGYSL | | Descriptor: | 1,2-ETHANEDIOL, ASP-PHE-GLU-ARG-GLU-GLY-TYR-SER-LEU, Beta-2-microglobulin, ... | | Authors: | Rizkallah, P.J, Tungatt, K, Sewell, A.K. | | Deposit date: | 2017-04-19 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Induction of influenza-specific local CD8 T-cells in the respiratory tract after aerosol delivery of vaccine antigen or virus in the Babraham inbred pig.
Plos Pathog., 14, 2018
|
|
6S96
 
 | |
6S9F
 
 | | Drosophila OTK, extracellular domains 3-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, Tyrosine-protein kinase-like otk | | Authors: | Rozbesky, D, Jones, E.Y. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.969 Å) | | Cite: | Drosophila OTK Is a Glycosaminoglycan-Binding Protein with High Conformational Flexibility.
Structure, 28, 2020
|
|
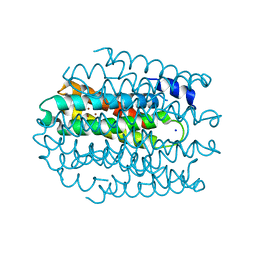
5NQM
 
 | |
6SC9
 
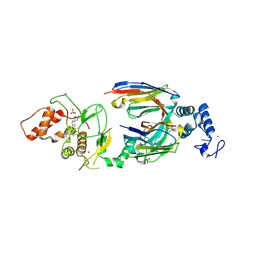 | | dAb3/HOIP-RBR-HOIPIN-8 | | Descriptor: | 2-[3-[2,6-bis(fluoranyl)-4-(1~{H}-pyrazol-4-yl)phenyl]-3-oxidanylidene-prop-1-enyl]-4-(1-methylpyrazol-4-yl)benzoic acid, CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Tsai, Y.-C.I, Johansson, H, House, D, Rittinger, K. | | Deposit date: | 2019-07-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Single-Domain Antibodies as Crystallization Chaperones to Enable Structure-Based Inhibitor Development for RBR E3 Ubiquitin Ligases.
Cell Chem Biol, 27, 2020
|
|
5OBZ
 
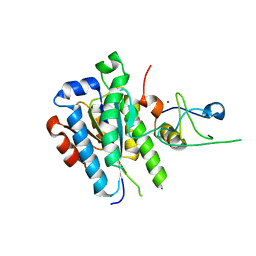 | | low resolution structure of the p34ct/p44ct minimal complex | | Descriptor: | Putative transcription factor, ZINC ION | | Authors: | Schoenwetter, E, Koelmel, W, Schmitt, D.R, Kuper, J, Kisker, C. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
6SFI
 
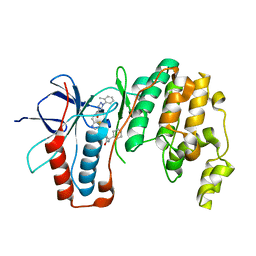 | | Crystal structure of p38 alpha in complex with compound 75 (MCP33) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]-2-[[[1-(2-methylphenyl)pyrazol-4-yl]carbonylamino]methyl]-1,3-thiazole-5-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
5OCC
 
 | |
5NZ1
 
 | |
5O08
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with Dephospho-coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, DEPHOSPHO COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0D
 
 | | Crystal structure of Phosphopantetheine adenylyltransferase from Mycobacterium abcessus in complex with 3-Phenoxymandelic acid (Fragment 4) | | Descriptor: | (2~{S})-2-oxidanyl-2-(3-phenoxyphenyl)ethanoic acid, Phosphopantetheine adenylyltransferase | | Authors: | Thomas, S.E, Kim, S.Y, Mendes, V, Blaszczyk, M, Blundell, T.L. | | Deposit date: | 2017-05-16 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structural Biology and the Design of New Therapeutics: From HIV and Cancer to Mycobacterial Infections: A Paper Dedicated to John Kendrew.
J. Mol. Biol., 429, 2017
|
|
5O0S
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with unreacted beta Cyclophellitol Cyclosulfate probe ME711 | | Descriptor: | (3~{a}~{S},4~{R},5~{S},6~{R},7~{R},7~{a}~{R})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydrobenzo[d][1,3,2]dioxathiole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-05-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
5NSC
 
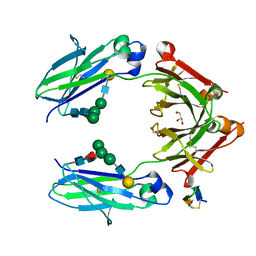 | | Fc DEKK heterodimer variant | | Descriptor: | Fc-III peptide, GLYCEROL, Putative uncharacterized protein DKFZp686C11235, ... | | Authors: | De Nardis, C, Hendriks, L.J.A, Poirier, E, Arvinte, T, Gros, P, Bakker, A.B.H, de Kruif, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-07-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A new approach for generating bispecific antibodies based on a common light chain format and the stable architecture of human immunoglobulin G1.
J. Biol. Chem., 292, 2017
|
|
6SAO
 
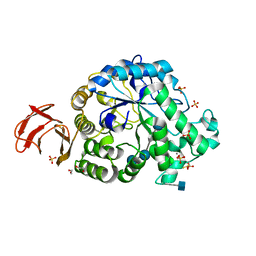 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
5NT1
 
 | |
5NTH
 
 | |
5NX2
 
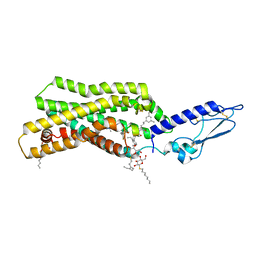 | | Crystal structure of thermostabilised full-length GLP-1R in complex with a truncated peptide agonist at 3.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucagon-like peptide 1 receptor, ... | | Authors: | Rappas, M, Jazayeri, A, Brown, A.J.H, Kean, J, Errey, J.C, Robertson, N, Fiez-Vandal, C, Andrews, S.P, Congreve, M, Bortolato, A, Mason, J.S, Baig, A.H, Teobald, I, Dore, A.S, Weir, M, Cooke, R.M, Marshall, F.H. | | Deposit date: | 2017-05-09 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal structure of the GLP-1 receptor bound to a peptide agonist.
Nature, 546, 2017
|
|
6SF6
 
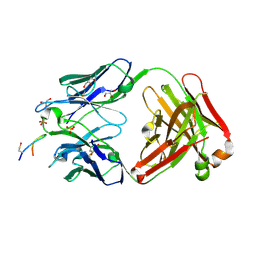 | |
6SEM
 
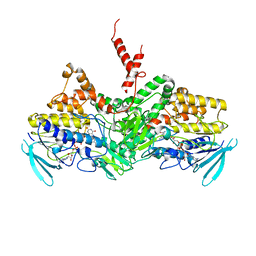 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 2 | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SFJ
 
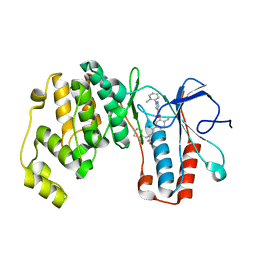 | | Crystal structure of p38 alpha in complex with compound 77 (MCP41) | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[5-[[(2~{S})-1-azanyl-4-cyclohexyl-1-oxidanylidene-butan-2-yl]carbamoyl]-2-methyl-phenyl]-1-(2-methylphenyl)pyrazole-4-carboxamide | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fast Iterative Synthetic Approach toward Identification of Novel Highly Selective p38 MAP Kinase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6SG2
 
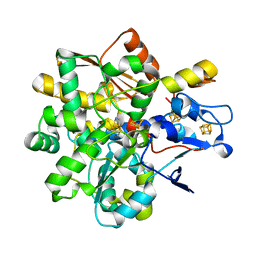 | | FeFe Hydrogenase from Desulfovibrio desulfuricans in Hinact state | | Descriptor: | HydB, IRON/SULFUR CLUSTER, Periplasmic [Fe] hydrogenase large subunit, ... | | Authors: | Galle, L.M, Span, I. | | Deposit date: | 2019-08-02 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Caught in the H inact : Crystal Structure and Spectroscopy Reveal a Sulfur Bound to the Active Site of an O 2 -stable State of [FeFe] Hydrogenase.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5NUN
 
 | | Engineered beta-lactoglobulin: variant F105L-L39A-M107F in complex with chlorpromazine (LG-LAF-CLP) | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Beta-lactoglobulin | | Authors: | Loch, J.I, Bonarek, P, Towrzydlo, M, Lazinskia, I, Szydlowska, J, Lewinski, K. | | Deposit date: | 2017-04-30 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The engineered beta-lactoglobulin with complementarity to the chlorpromazine chiral conformers.
Int. J. Biol. Macromol., 114, 2018
|
|
5O2U
 
 | | Llama VHH in complex with p24 | | Descriptor: | Capsid protein p24, VHH 59H10 | | Authors: | Caillat, C, Verrips, T, Weissenhorn, W. | | Deposit date: | 2017-05-22 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.
ACS Infect Dis, 3, 2017
|
|
