6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
4QXG
 
 | |
1DGD
 
 | |
2H6P
 
 | | Crystal structure of HLA-B*3501 presenting the human cytochrome P450 derived peptide, KPIVVLHGY | | Descriptor: | 9 mer peptide from Cytochrome P450, Beta-2-microglobulin, HLA-B35 | | Authors: | Archbold, J.K, Macdonald, W.A, Rossjohn, J. | | Deposit date: | 2006-05-31 | | Release date: | 2006-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alloreactivity between disparate cognate and allogeneic pMHC-I complexes is the result of highly focused, peptide-dependent structural mimicry
J.Biol.Chem., 281, 2006
|
|
1DGE
 
 | |
1Y5X
 
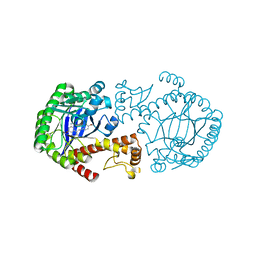 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methoxyphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHOXYPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
4HDS
 
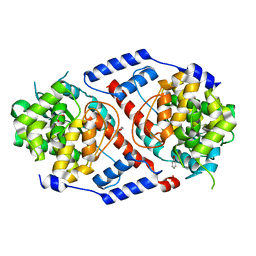 | | Crystal Structure of ArsAB in Complex with Phenol. | | Descriptor: | 1,2-ETHANEDIOL, ArsA, ArsB, ... | | Authors: | Newmister, S.A, Chan, C.H, Escalante-Semerena, J.C, Rayment, I. | | Deposit date: | 2012-10-02 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insights into the Function of the Nicotinate Mononucleotide:phenol/p-cresol Phosphoribosyltransferase (ArsAB) Enzyme from Sporomusa ovata.
Biochemistry, 51, 2012
|
|
1Y5W
 
 | | tRNA-guanine Transglycosylase (TGT) in complex with 6-Amino-4-[2-(4-methylphenyl)ethyl]-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-AMINO-4-[2-(4-METHYLPHENYL)ETHYL]-1,7-DIHYDRO-8H-IMIDAZO[4,5-G]QUINAZOLIN-8-ONE, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Stengl, B, Meyer, E.A, Heine, A, Brenk, R, Diederich, F, Klebe, G. | | Deposit date: | 2004-12-03 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of tRNA-guanine transglycosylase (TGT) in complex with novel and potent inhibitors unravel pronounced induced-fit adaptations and suggest dimer formation upon substrate binding
J.Mol.Biol., 370, 2007
|
|
1Y6Q
 
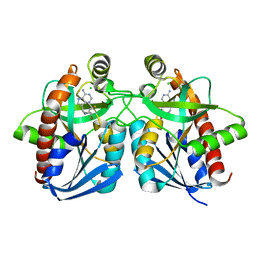 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
3RLA
 
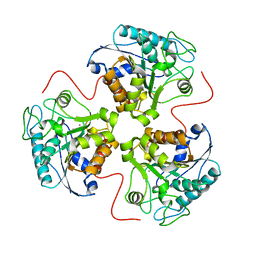 | |
1Y4T
 
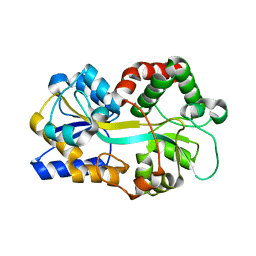 | | Ferric binding protein from Campylobacter jejuni | | Descriptor: | FE (III) ION, putative iron-uptake ABC transport system periplasmic iron-binding protein | | Authors: | Tom-Yew, S.A.L, Cui, D.T, Bekker, E.G, Murphy, M.E.P. | | Deposit date: | 2004-12-01 | | Release date: | 2005-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Anion-independent iron coordination by the Campylobacter jejuni ferric binding protein
J.Biol.Chem., 280, 2005
|
|
3PF3
 
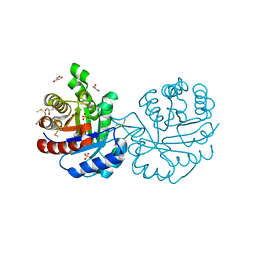 | | Crystal structure of a mutant (C202A) of Triosephosphate isomerase from Giardia lamblia derivatized with MMTS | | Descriptor: | CALCIUM ION, GLYCEROL, SULFATE ION, ... | | Authors: | Enriquez-Flores, S, Rodriguez-Romero, A, Hernandez-Santoyo, A, Reyes-Vivas, H. | | Deposit date: | 2010-10-27 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Determining the molecular mechanism of inactivation by chemical modification of triosephosphate isomerase from the human parasite Giardia lamblia: A study for antiparasitic drug design.
Proteins, 79, 2011
|
|
1HPJ
 
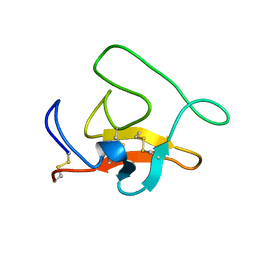 | |
2N9D
 
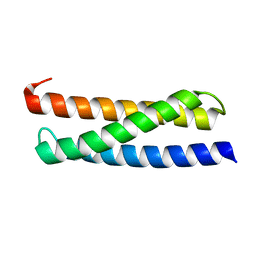 | |
2MWH
 
 | | NMR solution structure of ligand-free OAA | | Descriptor: | Anti-HIV lectin OAA | | Authors: | Lee, D, Carneiro, M.G, Koharudin, L.M, Griesinger, C, Gronenborn, A.M, Ban, D, Sabo, T, Trigo-Mourino, P, Mazur, A. | | Deposit date: | 2014-11-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Sampling of Glycan-Bound Conformers by the Anti-HIV Lectin Oscillatoria agardhii agglutinin in the Absence of Sugar.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5DAA
 
 | |
1I6Q
 
 | | Formation of a protein intermediate and its trapping by the simultaneous crystallization process: Crystal structure of an iron-saturated intermediate in the FE3+ binding pathway of camel lactoferrin at 2.7 resolution | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Khan, J.A, Kumar, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2001-03-03 | | Release date: | 2001-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Protein intermediate trapped by the simultaneous crystallization process. Crystal structure of an iron-saturated intermediate in the Fe3+ binding pathway of camel lactoferrin at 2.7 a resolution.
J.Biol.Chem., 276, 2001
|
|
3WIP
 
 | | Crystal structure of acetylcholine bound to Ls-AChBP | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholine-binding protein, ... | | Authors: | Olsen, J.A, Balle, T, Gajhede, M, Ahring, P.K, Kastrup, J.S. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition of the neurotransmitter acetylcholine by an acetylcholine binding protein reveals determinants of binding to nicotinic acetylcholine receptors
Plos One, 9, 2014
|
|
5DFP
 
 | | Crystal structure of PAK1 in complex with an inhibitor compound FRAX1036 | | Descriptor: | 6-[2-chloro-4-(6-methylpyrazin-2-yl)phenyl]-8-ethyl-2-{[2-(1-methylpiperidin-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidin-7(8H)-one, DIMETHYL SULFOXIDE, Serine/threonine-protein kinase PAK 1 | | Authors: | Maksimoska, J, Marmorstein, R, Wang, W. | | Deposit date: | 2015-08-27 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of Selective PAK1 Inhibitor G-5555: Improving Properties by Employing an Unorthodox Low-pK a Polar Moiety.
Acs Med.Chem.Lett., 6, 2015
|
|
4Z35
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | Descriptor: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | Authors: | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | Deposit date: | 2015-03-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
6Q1L
 
 | | Crystal structure of oxidized iodotyrosine deiodinase (IYD) bound to FMN and 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sun, Z, Kavran, J.M, Rokita, S.E. | | Deposit date: | 2019-08-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The minimal structure for iodotyrosine deiodinase function is defined by an outlier protein from the thermophilic bacterium Thermotoga neapolitana.
J.Biol.Chem., 297, 2021
|
|
7Y6Z
 
 | | RLGSGG-AtPRT6 UBR box (I222) | | Descriptor: | E3 ubiquitin-protein ligase PRT6, ZINC ION | | Authors: | Kim, L, Song, H.K. | | Deposit date: | 2022-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural analyses of plant PRT6-UBR box for Cys-Arg/N-degron pathway and insights into the plant submergence resistance
To Be Published
|
|
1LFO
 
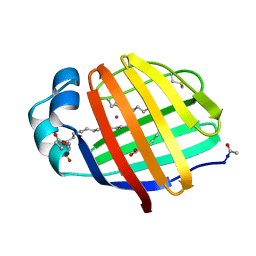 | | LIVER FATTY ACID BINDING PROTEIN-OLEATE COMPLEX | | Descriptor: | BUTENOIC ACID, LIVER FATTY ACID BINDING PROTEIN, OLEIC ACID, ... | | Authors: | Thompson, J, Winter, N, Terwey, D, Bratt, J, Banaszak, L. | | Deposit date: | 1996-12-09 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the liver fatty acid-binding protein. A complex with two bound oleates.
J.Biol.Chem., 272, 1997
|
|
1ZNV
 
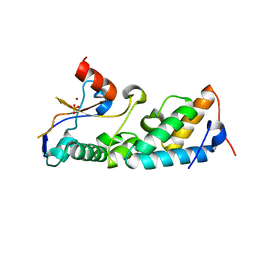 | | How a His-metal finger endonuclease ColE7 binds and cleaves DNA with a transition metal ion cofactor | | Descriptor: | Colicin E7, Colicin E7 immunity protein, NICKEL (II) ION, ... | | Authors: | Doudeva, L.G, Huang, H, Hsia, K.C, Shi, Z, Li, C.L, Shen, Y, Yuan, H.S. | | Deposit date: | 2005-05-12 | | Release date: | 2006-03-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis and metal-dependent stability and activity studies of the ColE7 endonuclease domain in complex with DNA/Zn2+ or inhibitor/Ni2+
Protein Sci., 15, 2006
|
|
3DGB
 
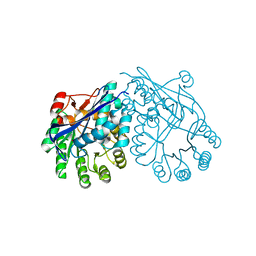 | | Crystal structure of muconate lactonizing enzyme from Pseudomonas Fluorescens complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
