9AYV
 
 | | HIV CH505/BG505 SOSIP.v8.1 Env in Complex with V1/V3 Epitope and Anti-Immune Complex pAbs from Rabbit 2474 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Rabbit Anti-Immune Complex pAb - Predicted Light Chain, ... | | Authors: | Brown, S, Antanasijevic, A, Ward, A.B. | | Deposit date: | 2024-03-08 | | Release date: | 2025-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Anti-immune complex antibodies are elicited during repeated immunization with HIV Env immunogens.
Sci Immunol, 10, 2025
|
|
3A4N
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
9IH8
 
 | | HSV-2 postfusion glycoprotein B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein B | | Authors: | Vollmer, B, Mulvaney, T, Ebel, H, Nentwig, J, Gruenewald, K. | | Deposit date: | 2025-02-20 | | Release date: | 2025-09-03 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | A nanobody specific to prefusion glycoprotein B neutralizes HSV-1 and HSV-2.
Nature, 2025
|
|
7TN5
 
 | |
5XZR
 
 | | The atomic structure of SHP2 E76A mutant in complex with allosteric inhibitor 9b | | Descriptor: | 4-(3-phenylphenyl)-N-(2,2,6,6-tetramethylpiperidin-4-yl)-1,3-thiazol-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Li, D, Xie, J, Zhu, J, Liu, C. | | Deposit date: | 2017-07-13 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric Inhibitors of SHP2 with Therapeutic Potential for Cancer Treatment.
J. Med. Chem., 60, 2017
|
|
6KRW
 
 | | Crystal Structure of AtPTP1 at 1.4 angstrom | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, IODIDE ION, ... | | Authors: | Zhao, Y.Y, Luo, Z.P, Wang, J, Wu, J.W. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of AtPTP1 at 1.4 Angstroms
To Be Published
|
|
9H9D
 
 | | Protein kinase CK2 catalytic subunit alpha (CSNK2A1 gene product) in complex the the indenoindole-type inhibitor MC11 | | Descriptor: | 1,2,3,4-tetrakis(bromanyl)-5-propan-2-yl-7,8-dihydro-6~{H}-indeno[1,2-b]indole-9,10-dione, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Niefind, K, Lindenblatt, D, Werner, C. | | Deposit date: | 2024-10-30 | | Release date: | 2025-08-27 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Exploring the biological potential of the brominated indenoindole MC11 and its interaction with protein kinase CK2.
Biol.Chem., 406, 2025
|
|
7TN6
 
 | |
6RCT
 
 | | Crystal structure of CLK3 in complex with T3-CLK | | Descriptor: | 1,2-ETHANEDIOL, 4-[2-methyl-1-(4-methylpiperazin-1-yl)-1-oxidanylidene-propan-2-yl]-~{N}-(6-pyridin-4-ylimidazo[1,2-a]pyridin-2-yl)benzamide, Dual specificity protein kinase CLK3 | | Authors: | Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-04-11 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of CLK3 in complex with TP003
To Be Published
|
|
8IB6
 
 | | Respiratory complex Membrane domain of CI, focus-refined of type IA, Wild type mouse under cold temperature | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
8IAQ
 
 | | Respiratory complex Membrane domain of CI, focused map of type I, Wild type mouse under thermoneutral temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-09 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
5D25
 
 | | First bromodomain of BRD4 bound to inhibitor XD27 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 4-acetyl-N-[5-(diethylsulfamoyl)-2-hydroxyphenyl]-3,5-dimethyl-1H-pyrrole-2-carboxamide, Bromodomain-containing protein 4, ... | | Authors: | Wohlwend, D, Huegle, M, Gerhardt, S. | | Deposit date: | 2015-08-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 4-Acyl Pyrrole Derivatives Yield Novel Vectors for Designing Inhibitors of the Acetyl-Lysine Recognition Site of BRD4(1).
J.Med.Chem., 59, 2016
|
|
5V0A
 
 | | Crystal structure of human exonuclease 1 Exo1 (D225A) in complex with 5' recessed-end DNA (rVIII) | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*CP*GP*CP*TP*AP*GP*TP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(P*CP*GP*AP*CP*TP*AP*GP*CP*G)-3'), ... | | Authors: | Shi, Y, Beese, L.S. | | Deposit date: | 2017-02-28 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Interplay of catalysis, fidelity, threading, and processivity in the exo- and endonucleolytic reactions of human exonuclease I.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IC4
 
 | | Respiratory complex Membrane domain of CI, focus-refined of type I, PERK -/- mouse under cold temperature | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
8IBB
 
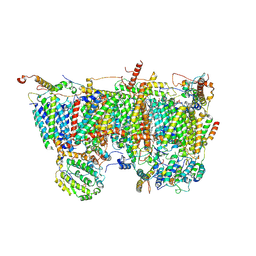 | | Respiratory complex Membrane domain of CI, focus-refined of type IB, Wild type mouse under cold temperature | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
8IBF
 
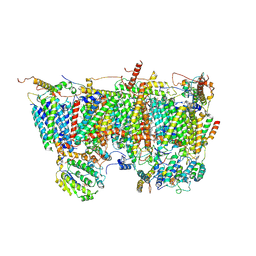 | | Respiratory complex Membrane domain of CI, focus-refined of type II, Wild type mouse under cold temperature | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, Acyl carrier protein, ... | | Authors: | Shin, Y.-C, Liao, M. | | Deposit date: | 2023-02-10 | | Release date: | 2024-09-18 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of respiratory complex adaptation to cold temperatures.
Cell, 187, 2024
|
|
3IEI
 
 | |
5Y6T
 
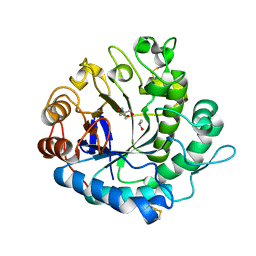 | | Crystal structure of endo-1,4-beta-mannanase from Eisenia fetida | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ISOPROPYL ALCOHOL, endo-1,4-beta-mannanase | | Authors: | Hirano, Y, Ueda, M, Tamada, T. | | Deposit date: | 2017-08-15 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gene cloning, expression, and X-ray crystallographic analysis of a beta-mannanase from Eisenia fetida.
Enzyme.Microb.Technol., 117, 2018
|
|
6ET5
 
 | | Reaction centre light harvesting complex 1 from Blc. virids | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Qian, P, Siebert, C.A, Canniffe, D.P, Wang, P, Hunter, C.N. | | Deposit date: | 2017-10-25 | | Release date: | 2018-04-11 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Cryo-EM structure of the Blastochloris viridis LH1-RC complex at 2.9 angstrom.
Nature, 556, 2018
|
|
1L7A
 
 | |
5D5V
 
 | | Crystal structure of human Hsf1 with Satellite III repeat DNA | | Descriptor: | DNA, Heat shock factor protein 1, MAGNESIUM ION | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5HGI
 
 | | Crystal structure of apo human IRE1 alpha | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BETA-MERCAPTOETHANOL, CESIUM ION, ... | | Authors: | Feldman, H.C, Tong, M, Wang, L, Meza-Acevedo, R, Gobillot, T.A, Gliedt, J.M, Hari, S.B, Mitra, A.K, Backes, B.J, Papa, F.R, Seeliger, M.A, Maly, D.J. | | Deposit date: | 2016-01-08 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.584 Å) | | Cite: | Structural and Functional Analysis of the Allosteric Inhibition of IRE1 alpha with ATP-Competitive Ligands.
Acs Chem.Biol., 11, 2016
|
|
7C6T
 
 | | Crystal structure of beta-glycosides-binding protein (W177X) of ABC transporter in a closed state bound to laminaritriose (Form I) | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
1VIS
 
 | | Crystal structure of mevalonate kinase | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Mevalonate kinase | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1LIF
 
 | |
