1P63
 
 | | Human Acidic Fibroblast Growth Factor. 140 Amino Acid Form with Amino Terminal His Tag and Leu111 Replaced with Ile (L111I) | | Descriptor: | ACIDIC FIBROBLAST GROWTH FACTOR, FORMIC ACID, SULFATE ION | | Authors: | Brych, S.R, Kim, J, Logan, T.M, Blaber, M. | | Deposit date: | 2003-04-28 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein
superfold
Protein Sci., 12, 2003
|
|
1IC7
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD32A99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1IC5
 
 | | CRYSTAL STRUCTURE OF HYHEL-10 FV MUTANT(HD99A)-HEN LYSOZYME COMPLEX | | Descriptor: | IGG1 FAB CHAIN H, LYSOZYME BINDING IG KAPPA CHAIN, LYSOZYME C | | Authors: | Shiroishi, M, Yokota, A, Tsumoto, K, Kondo, H, Nishimiya, Y, Horii, K, Matsushima, M, Ogasahara, K, Yutani, K, Kumagai, I. | | Deposit date: | 2001-03-30 | | Release date: | 2001-07-18 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence for entropic contribution of salt bridge formation to a protein antigen-antibody interaction: the case of hen lysozyme-HyHEL-10 Fv complex.
J.Biol.Chem., 276, 2001
|
|
1JT4
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND VAL 109 REPLACED BY LEU (V109L) | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1JT7
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND LEU 44 REPLACED BY PHE AND LEU 73 REPLACED BY VAL AND VAL 109 REPLACED BY LEU (L44F/L73V/V109L) | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1PWE
 
 | | Rat Liver L-Serine Dehydratase Apo Enzyme | | Descriptor: | L-serine dehydratase | | Authors: | Yamada, T, Komoto, J, Takata, Y, Ogawa, H, Takusagawa, F. | | Deposit date: | 2003-07-01 | | Release date: | 2003-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of serine dehydratase from rat liver.
Biochemistry, 42, 2003
|
|
1LLO
 
 | | HEVAMINE A (A PLANT ENDOCHITINASE/LYSOZYME) COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, Hevamine-A | | Authors: | Terwisscha Van Scheltinga, A.C, Armand, S, Kalk, K.H, Isogai, A, Henrissat, B, Dijkstra, B.W. | | Deposit date: | 1995-11-08 | | Release date: | 1996-03-08 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis.
Biochemistry, 34, 1995
|
|
1KY5
 
 | | D244E mutant S-Adenosylhomocysteine hydrolase refined with noncrystallographic restraints | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3'-OXO-ADENOSINE, S-adenosylhomocysteine hydrolase | | Authors: | Takata, Y, Takusagawa, F. | | Deposit date: | 2002-02-03 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Catalytic Mechanism of S-adenosylhomocysteine hydrolase. Site-directed
mutagenesis of Asp-130, Lys-185, Asp-189, and Asn-190.
J.Biol.Chem., 277, 2002
|
|
1JTC
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag AND LEU 44 REPLACED BY PHE (L44F) | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-20 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1K0U
 
 | | Inhibition of S-adenosylhomocysteine Hydrolase by "acyclic sugar" Adenosine Analogue D-eritadenine | | Descriptor: | D-ERITADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYL-L-HOMOCYSTEINE HYDROLASE | | Authors: | Takusagawa, F, Huang, Y, Komoto, J, Takata, Y, Gomi, T, Ogawa, H, Fujioka, M, Powell, D. | | Deposit date: | 2001-09-20 | | Release date: | 2001-10-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of S-adenosylhomocysteine hydrolase by acyclic sugar adenosine analogue D-eritadenine. Crystal structure of S-adenosylhomocysteine hydrolase complexed with D-eritadenine.
J.Biol.Chem., 277, 2002
|
|
1LTL
 
 | | THE DODECAMER STRUCTURE OF MCM FROM ARCHAEAL M. THERMOAUTOTROPHICUM | | Descriptor: | DNA replication initiator (Cdc21/Cdc54), ZINC ION | | Authors: | Fletcher, R.J, Bishop, B.E, Leon, R.P, Sclafani, R.A, Ogata, C.M, Chen, X.S. | | Deposit date: | 2002-05-20 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure and function of MCM from archaeal M. Thermoautotrophicum
Nat.Struct.Biol., 10, 2003
|
|
1J0O
 
 | | High Resolution Crystal Structure of the wild type Tetraheme Cytochrome c3 from Desulfovibrio vulgaris Miyazaki F | | Descriptor: | Cytochrome c3, ETHANOL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ozawa, K, Yasukawa, F, Kumagai, J, Ohmura, T, Cusanvich, M.A, Tomimoto, Y, Ogata, H, Higuchi, Y, Akutsu, H. | | Deposit date: | 2002-11-19 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Role of the aromatic ring of Tyr43 in tetraheme cytochrome c(3) from Desulfovibrio vulgaris Miyazaki F.
Biophys.J., 85, 2003
|
|
1IS4
 
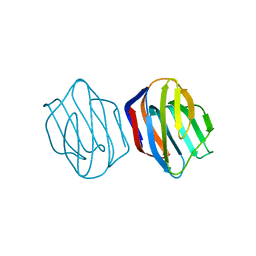 | | LACTOSE-LIGANDED CONGERIN II | | Descriptor: | CONGERIN II, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of a Conger Eel Galectin (Congerin II) at 1.45 A Resolution: Implication for the Accelerated Evolution of a New Ligand-Binding Site Following Gene Duplication
J.Mol.Biol., 321, 2002
|
|
3S3E
 
 | |
1JQZ
 
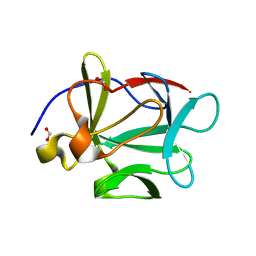 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag. | | Descriptor: | FORMIC ACID, acidic fibroblast growth factor | | Authors: | Brych, S.R, Blaber, S.I, Logan, T.M, Blaber, M. | | Deposit date: | 2001-08-09 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and stability effects of mutations designed to increase the primary sequence symmetry within the core
region of a beta-trefoil.
Protein Sci., 10, 2001
|
|
1OYX
 
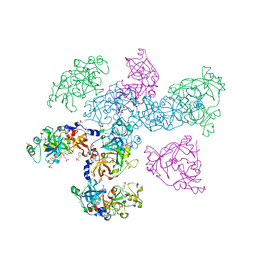 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (SELENO-MET) AT 1.85 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1OZ2
 
 | | CRYSTAL STRUCTURE OF 3-MBT REPEATS OF LETHAL (3) MALIGNANT BRAIN TUMOR (NATIVE-II) AT 1.55 ANGSTROM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lethal(3)malignant brain tumor-like protein, SULFATE ION | | Authors: | Wang, W.K, Tereshko, V, Boccuni, P, MacGrogan, D, Nimer, S.D, Patel, D.J. | | Deposit date: | 2003-04-07 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Malignant brain tumor repeats: a three-leaved propeller architecture with ligand/peptide binding pockets.
Structure, 11, 2003
|
|
1IS6
 
 | | MES-Liganded Congerin II | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Congerin II | | Authors: | Shirai, T, Matsui, Y, Shionyu-Mitsuyama, C, Yamane, T, Kamiya, H, Ishii, C, Ogawa, T, Muramoto, K. | | Deposit date: | 2001-11-12 | | Release date: | 2002-09-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a conger eel galectin (congerin II) at 1.45 A resolution: Implication for the accelerated evolution of a new ligand-binding site following gene duplication
J.MOL.BIOL., 321, 2002
|
|
1M4R
 
 | | CRYSTAL STRUCTURE OF RECOMBINANT HUMAN INTERLEUKIN-22 | | Descriptor: | Interleukin-22 | | Authors: | Nagem, R.A.P, Colau, D, Dumoutier, L, Renauld, J.-C, Ogata, C, Polikarpov, I. | | Deposit date: | 2002-07-03 | | Release date: | 2003-07-07 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Recombinant Human Interleukin-22
Structure, 10, 2002
|
|
1M16
 
 | | Human Acidic Fibroblast Growth Factor. 141 Amino Acid Form with Amino Terminal His Tag and Leu 44 Replaced with Phe (L44F), Leu 73 Replaced with Val (L73V), Val 109 Replaced with Leu (V109L) and Cys 117 Replaced with Val (C117V). | | Descriptor: | FORMIC ACID, SULFATE ION, acidic fibroblast growth factor | | Authors: | Brych, S.R, Kim, J, Spielmann, G.L, Logan, T.M, Blaber, M. | | Deposit date: | 2002-06-17 | | Release date: | 2003-08-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accommodation of a highly symmetric core within a symmetric protein superfold
Protein Sci., 12, 2003
|
|
1QW1
 
 | | Solution Structure of the C-Terminal Domain of DtxR residues 110-226 | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-29 | | Release date: | 2005-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
1QVP
 
 | | C terminal SH3-like domain from Diphtheria toxin Repressor residues 144-226. | | Descriptor: | Diphtheria toxin repressor | | Authors: | Wylie, G.P, Rangachari, V, Bienkiewicz, E.A, Marin, V, Bhattacharya, N, Love, J.F, Murphy, J.R, Logan, T.M. | | Deposit date: | 2003-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Prolylpeptide binding by the prokaryotic SH3-like domain of the diphtheria toxin repressor: a regulatory switch.
Biochemistry, 44, 2005
|
|
2ZBG
 
 | | Calcium pump crystal structure with bound AlF4 and TG in the absence of calcium | | Descriptor: | MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, 3ABETA, ... | | Authors: | Toyoshima, C, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2J5S
 
 | | Structural of ABDH, a beta-diketone hydrolase from the Cyanobacterium Anabaena sp. PCC 7120 bound to (S)-3-oxocyclohexyl acetic acid | | Descriptor: | (S)-CYCLOHEXANONE-2-ACETATE, BETA-DIKETONE HYDROLASE, NICKEL (II) ION | | Authors: | Bennett, J.P, Whittingham, J.L, Brzozowski, A.M, Leonard, P.M, Grogan, G. | | Deposit date: | 2006-09-19 | | Release date: | 2007-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Characterisation of a Beta Diketone Hydrolase from the Cyanobacterium Anabaena Sp. Pcc 7120 in Native and Product Bound Forms, a Coenzyme A-Independent Member of the Crotonase Suprafamily
Biochemistry, 46, 2007
|
|
2J5I
 
 | | Crystal Structure of Hydroxycinnamoyl-CoA Hydratase-Lyase | | Descriptor: | P-HYDROXYCINNAMOYL COA HYDRATASE/LYASE | | Authors: | Leonard, P.M, Brzozowski, A.M, Lebedev, A, Marshall, C.M, Smith, D.J, Verma, C.S, Walton, N.J, Grogan, G. | | Deposit date: | 2006-09-18 | | Release date: | 2006-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The 1.8 A Resolution Structure of Hydroxycinnamoyl- Coenzyme a Hydratase-Lyase (Hchl) from Pseudomonas Fluorescens, an Enzyme that Catalyses the Transformation of Feruloyl-Coenzyme a to Vanillin.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
