3THI
 
 | |
8EHG
 
 | | Rabbit muscle aldolase determined using single-particle cryo-EM with Apollo camera. | | Descriptor: | Fructose-bisphosphate aldolase A | | Authors: | Peng, R, Fu, X, Mendez, J.H, Randolph, P.H, Bammes, B, Stagg, S.M. | | Deposit date: | 2022-09-14 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Characterizing the resolution and throughput of the Apollo direct electron detector.
J Struct Biol X, 7, 2023
|
|
8EGD
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | 5-(4-methoxyphenyl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGQ
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2S)-2-ethyl-4-{[(2'M)-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl}-3,4-dihydro-2H-pyrido[3,2-b][1,4]oxazine, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.955 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGF
 
 | | Branched chain ketoacid dehydrogenase kinase in complex with inhibitor | | Descriptor: | (5P)-5-(4'-methyl[1,1'-biphenyl]-2-yl)-1H-tetrazole, ADENOSINE-5'-DIPHOSPHATE, POTASSIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-12 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8EGU
 
 | | Branched chain ketoacid dehydrogenase kinase complexes | | Descriptor: | (2~{S})-3-methyl-2-[pentanoyl-[[4-[2-(2~{H}-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]amino]butanoic acid, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.923 Å) | | Cite: | Structural studies identify angiotensin II receptor blocker-like compounds as branched-chain ketoacid dehydrogenase kinase inhibitors.
J.Biol.Chem., 299, 2023
|
|
8F5F
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2P)-2-[(4P)-4-{6-[(1-ethylcyclopropyl)methoxy]pyridin-3-yl}-1,3-thiazol-2-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.149 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F5J
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | 3-chloro-5-fluorothieno[3,2-b]thiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-14 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F5S
 
 | | human branched chain ketoacid dehydrogenase kinase in complex with inhibitors | | Descriptor: | (2M)-2-[2-(4-methylphenyl)-1,3-thiazol-4-yl]benzoic acid, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, S, Roth Flach, R, Bollinger, E, Filipski, K. | | Deposit date: | 2022-11-15 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.793 Å) | | Cite: | Small molecule branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors with opposing effects on BDK protein levels.
Nat Commun, 14, 2023
|
|
8F4Q
 
 | | rat branched chain ketoacid dehydrogenase kinase in complex with inhibtors | | Descriptor: | 3-chloro-5-fluorothieno[3,2-b]thiophene-2-carboxylic acid, ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Liu, S. | | Deposit date: | 2022-11-11 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Discovery of branched-chain ketoacid dehydrogenase kinase (BDK) inhibitors acting as stabilizers or destabilizers
To Be Published
|
|
1Q90
 
 | | Structure of the cytochrome b6f (plastohydroquinone : plastocyanin oxidoreductase) from Chlamydomonas reinhardtii | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 8-HYDROXY-5,7-DIMETHOXY-3-METHYL-2-TRIDECYL-4H-CHROMEN-4-ONE, ... | | Authors: | Stroebel, D, Choquet, Y, Popot, J.-L, Picot, D. | | Deposit date: | 2003-08-22 | | Release date: | 2003-12-09 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An Atypical Haem in the Cytochrome B6F Complex
Nature, 426, 2003
|
|
5MJI
 
 | | Crystal Structure of RosB with bound intermediate OHC-RP (8-demethyl-8-formylriboflavin-5'-phosphate) | | Descriptor: | BRAMP domain protein, [(2~{S},3~{R},4~{R})-5-[8-methanoyl-7-methyl-2,4-bis(oxidanylidene)-1~{H}-benzo[g]pteridin-10-ium-10-yl]-2,3,4-tris(oxidanyl)pentyl] dihydrogen phosphate | | Authors: | Konjik, V, Bruenle, S, Demmer, U, Vanselow, A, Sandhoff, R, Mack, M, Ermler, U. | | Deposit date: | 2016-12-01 | | Release date: | 2016-12-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of RosB: Insights into the Reaction Mechanism of the First Member of a Family of Flavodoxin-like Enzymes.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8QPL
 
 | | F420-Dependent Methylene-Tetrahydromethanopterin Reductase with F420 from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase, COENZYME F420 | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QPM
 
 | | Structure of methylene-tetrahydromethanopterin reductase from Methanocaldococcus jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
8QPJ
 
 | |
8QQ8
 
 | | Crystal Structure of F420-dependent Methylene-Tetrahydromethanopterin Reductase Mutant E6Q from Methanocaldococcus Jannaschii | | Descriptor: | 5,10-methylenetetrahydromethanopterin reductase | | Authors: | Gehl, M, Demmer, U, Ermler, U, Shima, S. | | Deposit date: | 2023-10-04 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutational and structural studies of ( beta alpha ) 8 -barrel fold methylene-tetrahydropterin reductases utilizing a common catalytic mechanism.
Protein Sci., 33, 2024
|
|
4G2E
 
 | |
4GQC
 
 | | Crystal Structure of Aeropyrum pernix Peroxiredoxin Q Enzyme in Fully-Folded and Locally-Unfolded Conformations | | Descriptor: | DITHIANE DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Perkins, A, Karplus, P.A, Gretes, M.C, Nelson, K.J, Poole, L.B. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the Active Site Helix-to-Strand Conversion of CxxxxC Peroxiredoxin Q Enzymes.
Biochemistry, 51, 2012
|
|
4GQF
 
 | | Aeropyrum pernix Peroxiredoxin Q Enzyme in the Locally Unfolded Conformation | | Descriptor: | GLYCEROL, SULFATE ION, Thiol peroxidase | | Authors: | Perkins, A, Karplus, P.A, Gretes, M.C, Nelson, K.J, Poole, L.B. | | Deposit date: | 2012-08-22 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping the Active Site Helix-to-Strand Conversion of CxxxxC Peroxiredoxin Q Enzymes.
Biochemistry, 51, 2012
|
|
6I33
 
 | | Crystal structure of human glycine decarboxylase (P-protein) | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, Glycine dehydrogenase (decarboxylating), ... | | Authors: | Van Laer, B, Kapp, U, Leonard, G, Mueller-Dieckmann, C. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights in human glycine decarboxylase and comparison with the Neanderthal variant
To Be Published
|
|
6I35
 
 | | Crystal structure of human glycine decarboxylase (P-protein) bound with pyridoxyl-glycine-5'-monophosphate | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Van Laer, B, Kapp, U, Leonard, G, Mueller-Dieckmann, C. | | Deposit date: | 2018-11-05 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights in human glycine decarboxylase and comparison with the Neanderthal variant
To Be Published
|
|
6FAH
 
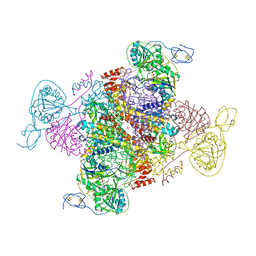 | | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction | | Descriptor: | Caffeyl-CoA reductase-Etf complex subunit CarC, Caffeyl-CoA reductase-Etf complex subunit CarD, Caffeyl-CoA reductase-Etf complex subunit CarE, ... | | Authors: | Demmer, J.K, Bertsch, J, Oeppinger, C, Wohlers, H, Kayastha, K, Demmer, U, Ermler, U, Mueller, V. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction.
FEBS Lett., 592, 2018
|
|
2A8X
 
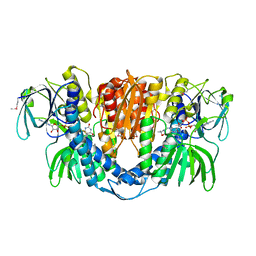 | | Crystal Structure of Lipoamide Dehydrogenase from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Rajashankar, K.R, Bryk, R, Kniewel, R, Buglino, J.A, Nathan, C.F, Lima, C.D. | | Deposit date: | 2005-07-10 | | Release date: | 2005-08-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and functional analysis of lipoamide dehydrogenase from Mycobacterium tuberculosis
J.Biol.Chem., 280, 2005
|
|
2XT6
 
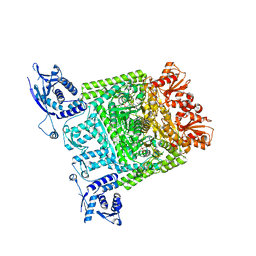 | | Crystal structure of Mycobacterium smegmatis alpha-ketoglutarate decarboxylase homodimer (orthorhombic form) | | Descriptor: | 2-OXOGLUTARATE DECARBOXYLASE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Bellinzoni, M, Wehenkel, A.M, O'Hare, H.M, Alzari, P.M. | | Deposit date: | 2010-10-05 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Functional Plasticity and Allosteric Regulation of Alpha-Ketoglutarate Decarboxylase in Central Mycobacterial Metabolism.
Chem.Biol., 18, 2011
|
|
4M52
 
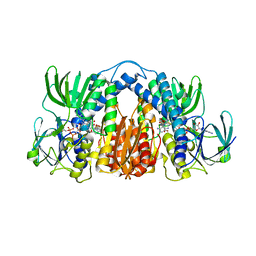 | | Structure of Mtb Lpd bound to SL827 | | Descriptor: | Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, N~2~-[(2-amino-5-bromopyridin-3-yl)sulfonyl]-N-(4-methoxyphenyl)-N~2~-methylglycinamide | | Authors: | Lima, C.D. | | Deposit date: | 2013-08-07 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipoamide channel-binding sulfonamides selectively inhibit mycobacterial lipoamide dehydrogenase.
Biochemistry, 52, 2013
|
|
