1BSX
 
 | | STRUCTURE AND SPECIFICITY OF NUCLEAR RECEPTOR-COACTIVATOR INTERACTIONS | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, PROTEIN (GRIP1), PROTEIN (THYROID HORMONE RECEPTOR BETA) | | Authors: | Wagner, R.L, Darimont, B.D, Apriletti, J.W, Stallcup, M.R, Kushner, P.J, Baxter, J.D, Fletterick, R.J, Yamamoto, K.R. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure and specificity of nuclear receptor-coactivator interactions.
Genes Dev., 12, 1998
|
|
3HBV
 
 | | PrtC methionine mutants: M226A in-house | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secreted protease C, ... | | Authors: | Oberholzer, A.E, Bumann, M, Hege, T, Russo, S, Baumann, U. | | Deposit date: | 2009-05-05 | | Release date: | 2009-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Metzincin's canonical methionine is responsible for the structural integrity of the zinc-binding site
Biol.Chem., 390, 2009
|
|
3U2P
 
 | | Crystal structure of N-terminal three extracellular domains of ErbB4/Her4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Bouyain, S, Elgenbrot, C, Leahy, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The ErbB4 extracellular region retains a tethered-like conformation in the absence of the tether.
Protein Sci., 21, 2012
|
|
2RMF
 
 | | Human Urocortin 1 | | Descriptor: | Urocortin | | Authors: | Grace, C.R.R, Perrin, M.H, Cantle, J.P, Vale, W.W, Rivier, J.E, Riek, R. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Common and divergent structural features of a series of corticotropin releasing factor-related peptides
J.Am.Chem.Soc., 129, 2007
|
|
2RMG
 
 | | Human Urocortin 2 | | Descriptor: | Urocortin-2 | | Authors: | Grace, C.R.R, Perrin, M.H, Cantle, J.P, Vale, W.W, Rivier, J.E, Riek, R. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Common and divergent structural features of a series of corticotropin releasing factor-related peptides
J.Am.Chem.Soc., 129, 2007
|
|
3JPV
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a pyrrolo[2,3-a]carbazole ligand | | Descriptor: | 1,10-dihydropyrrolo[2,3-a]carbazole-3-carbaldehyde, Peptide (PIMTIDE) ARKRRRHPSGPPTA, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Filippakopoulos, P, Bullock, A.N, Fedorov, O, Akue-Gedu, R, Rossignol, E, Azzaro, S, Bain, J, Cohen, P, Prudhomme, M, Moreau, P, Amizon, F, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-04 | | Release date: | 2009-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Synthesis, kinase inhibitory potencies, and in vitro antiproliferative evaluation of new pim kinase inhibitors.
J.Med.Chem., 52, 2009
|
|
3U8J
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3531 (1-(pyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(pyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
2H77
 
 | | Crystal structure of human TR alpha bound T3 in monoclinic space group | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, THRA protein | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Bleicher, L, Ambrosio, A.L.B, Figueira, A.C.M, Santos, M.A.M, Neto, M.O, Fischer, H, Togashi, H.F.M, Craievich, A.F, Garrat, R.C, Baxter, J.D, Webb, P, Polikarpov, I. | | Deposit date: | 2006-06-01 | | Release date: | 2006-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function.
J.Mol.Biol., 360, 2006
|
|
7EIP
 
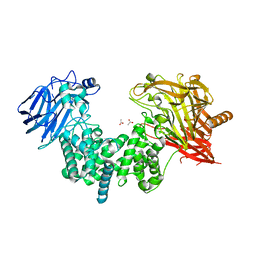 | | Crystal structure of ligand-free chondroitin ABC lyase I | | Descriptor: | ACETATE ION, Chondroitin sulfate ABC endolyase, MAGNESIUM ION | | Authors: | Takashima, M, Miyanaga, A, Eguchi, T. | | Deposit date: | 2021-03-31 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate specificity of Chondroitinase ABC I based on analyses of biochemical reactions and crystal structures in complex with disaccharides.
Glycobiology, 31, 2021
|
|
4KJV
 
 | | Crystal structure of XIAP-Bir2 with a bound spirocyclic benzoxazepinone inhibitor. | | Descriptor: | 6-methoxy-5-({(3S)-3-[(N-methyl-L-alanyl)amino]-4-oxo-2',3,3',4,5',6'-hexahydro-5H-spiro[1,5-benzoxazepine-2,4'-pyran]-5-yl}methyl)naphthalene-2-carboxylic acid, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Lukacs, C.M, Janson, C.A. | | Deposit date: | 2013-05-03 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of Benzodiazepinones as Selective Inhibitors of the X-Linked Inhibitor of Apoptosis Protein (XIAP) Second Baculovirus IAP Repeat (BIR2) Domain.
J.Med.Chem., 56, 2013
|
|
1K6L
 
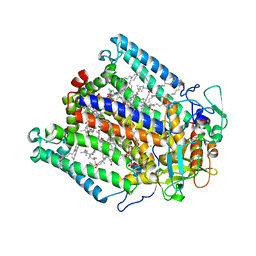 | | Photosynethetic Reaction Center from Rhodobacter sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Laible, P.D, Deng, Y.-L, Wong, T.N, Hanson, D.K, Schiffer, M. | | Deposit date: | 2001-10-16 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of a mutant photosynthetic reaction center shows unexpected changes in main chain orientations and quinone position.
Biochemistry, 41, 2002
|
|
2HEY
 
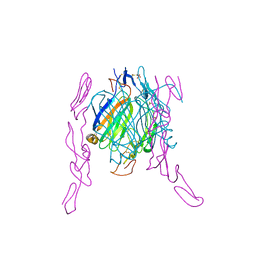 | | Crystal structure of murine OX40L bound to human OX40 | | Descriptor: | SULFATE ION, Tumor necrosis factor ligand superfamily member 4, Tumor necrosis factor receptor superfamily member 4 | | Authors: | Hymowitz, S.G. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Costimulatory OX40-OX40L Complex.
Structure, 14, 2006
|
|
1K6N
 
 | | E(L212)A,D(L213)A Double Mutant Structure of Photosynthetic Reaction Center from Rhodobacter Sphaeroides | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Laible, P.D, Deng, Y.-L, Wong, T.N, Hanson, D.K, Schiffer, M. | | Deposit date: | 2001-10-16 | | Release date: | 2002-08-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structure of a mutant photosynthetic reaction center shows unexpected changes in main chain orientations and quinone position.
Biochemistry, 41, 2002
|
|
2GWX
 
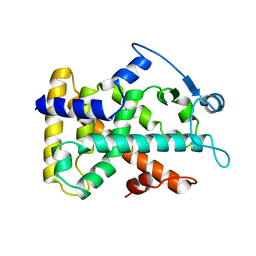 | | MOLECULAR RECOGNITION OF FATTY ACIDS BY PEROXISOME PROLIFERATOR-ACTIVATED RECEPTORS | | Descriptor: | PROTEIN (PPAR-DELTA) | | Authors: | Xu, H.E, Lambert, M.H, Montana, V.G, Park, D.J, Blanchard, S, Brown, P, Sternbach, D, Lehmann, J, Bruce, G.W, Willson, T.M, Kliewer, S.A, Milburn, M.V. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular recognition of fatty acids by peroxisome proliferator-activated receptors.
Mol.Cell, 3, 1999
|
|
3MA3
 
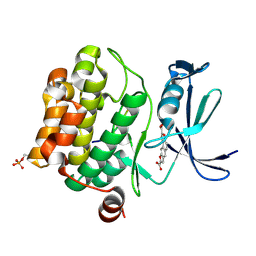 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a naphtho-difuran ligand | | Descriptor: | Pimtide, Proto-oncogene serine/threonine-protein kinase pim-1, naphtho[2,1-b:7,6-b']difuran-2,8-dicarboxylic acid | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Vollmar, M, von Delft, F, Cochet, C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New potent dual inhibitors of CK2 and Pim kinases: discovery and structural insights.
Faseb J., 24, 2010
|
|
4J3E
 
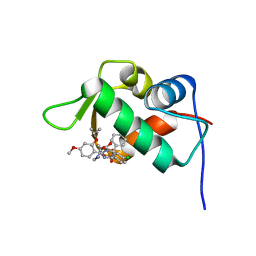 | | The 1.9A crystal structure of humanized Xenopus Mdm2 with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Graves, B.J, Lukacs, C.M, Kammlott, R.U, Crowther, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Discovery of RG7112: A Small-Molecule MDM2 Inhibitor in Clinical Development.
ACS Med Chem Lett, 4, 2013
|
|
3NFN
 
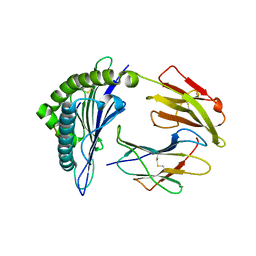 | | Recognition of peptide-MHC by a V-delta/V-beta TCR | | Descriptor: | 10-mer peptide from Protein Nef, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Shi, Y, Qi, J, Gao, F, Gao, G.F. | | Deposit date: | 2010-06-10 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | Plasticity of human CD8 alpha alpha binding to peptide-HLA-A*2402
Mol.Immunol., 48, 2011
|
|
3OZW
 
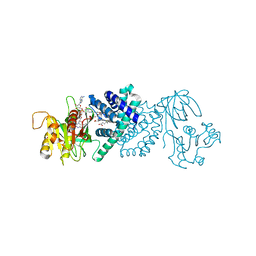 | | The Crystal Structure of flavohemoglobin from R. eutrophus in complex with ketoconazole | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, 1-acetyl-4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-imidazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazine, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Ermler, U, Baciou, L. | | Deposit date: | 2010-09-27 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Ralstonia eutropha Flavohemoglobin in Complex with Three Antibiotic Azole Compounds.
Biochemistry, 50, 2011
|
|
7CPP
 
 | |
4OQQ
 
 | |
2RMD
 
 | | Astressin-B | | Descriptor: | ASTRESSIN-B | | Authors: | Grace, C.R.R, Perrin, M.H, Cantle, J.P, Vale, W.W, Rivier, J.E, Riek, R. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-01 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Common and divergent structural features of a series of corticotropin releasing factor-related peptides
J.Am.Chem.Soc., 129, 2007
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
5U03
 
 | | Cryo-EM structure of the human CTP synthase filament | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CTP synthase 1, URIDINE 5'-TRIPHOSPHATE | | Authors: | Lynch, E.M, Kollman, J.M. | | Deposit date: | 2016-11-22 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Human CTP synthase filament structure reveals the active enzyme conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
2RM9
 
 | | Astressin2B | | Descriptor: | Astressin2B | | Authors: | Grace, C.R.R, Perrin, M.H, Cantle, J.P, Vale, W.W, Rivier, J.E, Riek, R. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-01 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Common and divergent structural features of a series of corticotropin releasing factor-related peptides
J.Am.Chem.Soc., 129, 2007
|
|
2RME
 
 | | Stressin | | Descriptor: | Stressin | | Authors: | Grace, C.R.R, Perrin, M.H, Cantle, J.P, Vale, W.W, Rivier, J.E, Riek, R. | | Deposit date: | 2007-10-16 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Common and divergent structural features of a series of corticotropin releasing factor-related peptides
J.Am.Chem.Soc., 129, 2007
|
|
