7B31
 
 | | MST3 in complex with compound MRIA9 | | Descriptor: | 1-[(5-azanyl-1,3-dioxan-2-yl)methyl]-3-[2-chloranyl-4-(3-fluoranylpyridin-2-yl)phenyl]-7-(methylamino)-1,6-naphthyridin-2-one, Serine/threonine-protein kinase 24 | | Authors: | Tesch, R, Rak, M, Joerger, A.C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-28 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of Selective Salt-Inducible Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
5DIE
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a trifluoro-substituted A-CD ring estrogen derivative (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol | | Descriptor: | (1S,3aR,5S,7aS)-7a-methyl-5-(2,3,5-trifluoro-4-hydroxyphenyl)octahydro-1H-inden-1-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-08-31 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7B6F
 
 | | GSK3-beta in complex with compound (S)-5c | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3~{S})-3-[(7-chloranyl-9~{H}-pyrimido[4,5-b]indol-4-yl)-methyl-amino]piperidin-1-yl]propanenitrile, Glycogen synthase kinase-3 beta, ... | | Authors: | Tesch, R, Andreev, S, Koch, P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | GSK3-beta in complex with compound (S)-5c
To be published
|
|
7XEY
 
 | | EDS1-PAD4 complexed with pRib-ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-beta-D-ribofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Huang, S, Jia, A, Xiao, Y. | | Deposit date: | 2022-03-31 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Identification and receptor mechanism of TIR-catalyzed small molecules in plant immunity.
Science, 377, 2022
|
|
4KTD
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate | | Descriptor: | GE136 Heavy Chain Fab, GE136 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Standfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7AY3
 
 | |
5VPS
 
 | | Crystal structure of an SDR from Burkholderia ambifaria in complex with NADPH with a TCEP adduct | | Descriptor: | 1,2-ETHANEDIOL, Short-chain dehydrogenase/reductase SDR, [(4~{S})-3-aminocarbonyl-1-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]piperidin-4-yl]-tris(3-hydroxy-3-oxopropyl)phosphanium | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-05-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Cautionary Tale of Using Tris(alkyl)phosphine Reducing Agents with NAD+-Dependent Enzymes.
Biochemistry, 2020
|
|
5KJR
 
 | |
3L2N
 
 | |
5VSE
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with inhibitor 17: N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-cyclopentyl-6,7-dimethoxy-N~2~-methyl-N~4~-(1-methylpiperidin-4-yl)quinazoline-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Babault, N, Xiong, Y, Liu, J, Jin, J. | | Deposit date: | 2017-05-11 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-activity relationship studies of G9a-like protein (GLP) inhibitors.
Bioorg. Med. Chem., 25, 2017
|
|
7SFC
 
 | | Human DNMT1(729-1600) Bound to Zebularine-Containing 12mer dsDNA and Inhibitor GSK3735967A | | Descriptor: | 1,2-ETHANEDIOL, DNA (12-MER), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Horton, J.R, Pathuri, S, Cheng, X. | | Deposit date: | 2021-10-03 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural characterization of dicyanopyridine containing DNMT1-selective, non-nucleoside inhibitors.
Structure, 30, 2022
|
|
5VSV
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P225 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, {2-chloro-5-[({2-[3-(prop-1-en-2-yl)phenyl]propan-2-yl}carbamoyl)amino]phenoxy}acetic acid | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-12 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P225
To Be Published
|
|
3BWU
 
 | | Crystal structure of the ternary complex of FimD (N-Terminal Domain, FimDN) with FimC and the N-terminally truncated pilus subunit FimF (FimFt) | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Eidam, O, Grutter, M.G, Capitani, G. | | Deposit date: | 2008-01-10 | | Release date: | 2008-03-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD
Febs Lett., 582, 2008
|
|
1US4
 
 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E, Takahashi, H. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
4Q1J
 
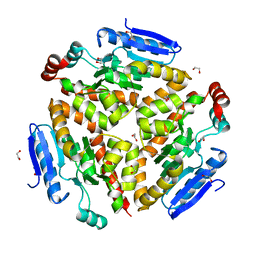 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
8IUF
 
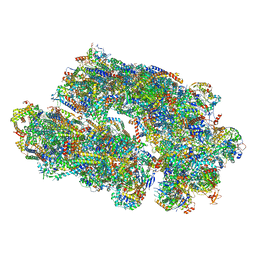 | | Cryo-EM structure of Euglena gracilis super-complex I+III2+IV, composite | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Tian, H.T, He, Z.X, Hu, Y.Q, Zhou, L. | | Deposit date: | 2023-03-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Euglena's atypical respiratory chain adapts to the discoidal cristae and flexible metabolism.
Nat Commun, 15, 2024
|
|
3ZXJ
 
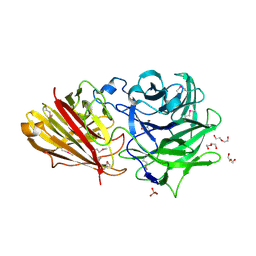 | | Engineering the active site of a GH43 glycoside hydrolase generates a biotechnologically significant enzyme that displays both endo- xylanase and exo-arabinofuranosidase activity | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, HIAXHD3, ... | | Authors: | McKee, L.S, Pena, M.J, Rogowski, A, Jackson, A, Lewis, R.J, York, W.S, Krogh, K.B.R.M, Vikso-Nielsen, A, Skjot, M, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-08-11 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Introducing Endo-Xylanase Activity Into an Exo-Acting Arabinofuranosidase that Targets Side Chains.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6FM7
 
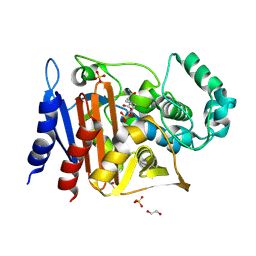 | | Crystal structure of the class C beta-lactamase TRU-1 from Aeromonas enteropelogenes in complex with avibactam | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Di Pisa, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2018-01-30 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Atomic-Resolution Structure of a Class C beta-Lactamase and Its Complex with Avibactam.
ChemMedChem, 13, 2018
|
|
1US5
 
 | | PUTATIVE GLUR0 LIGAND BINDING CORE WITH L-GLUTAMATE | | Descriptor: | 1,2-ETHANEDIOL, GLUTAMIC ACID, PUTATIVE GLUR0 LIGAND BINDING CORE | | Authors: | Tahirov, T.H, Inagaki, E. | | Deposit date: | 2003-11-18 | | Release date: | 2003-11-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Thermus Thermophilus Putative Periplasmic Glutamate/Glutamine-Binding Protein
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5ALZ
 
 | | ligand complex structure of soluble epoxide hydrolase | | Descriptor: | 1-(1-adamantyl)-3-(1-methylsulfonylpiperidin-4-yl)urea, BIFUNCTIONAL EPOXIDE HYDROLASE 2, SULFATE ION | | Authors: | Oster, L, Tapani, S, Xue, Y, Kack, H. | | Deposit date: | 2015-03-08 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Successful Generation of Structural Information for Fragment-Based Drug Discovery.
Drug Discov Today, 20, 2015
|
|
6G2M
 
 | | Crystal structure of human mitochondrial 5'(3')-deoxyribonucleotidase in complex with the inhibitor PB-PAU | | Descriptor: | 1,2-ETHANEDIOL, 5'(3')-deoxyribonucleotidase, mitochondrial, ... | | Authors: | Pachl, P, Rezacova, P, Brynda, J. | | Deposit date: | 2018-03-23 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure-based optimization of bisphosphonate nucleoside inhibitors of human 5'(3')-deoxyribonucleotidases
Eur.J.Org.Chem., 2018
|
|
8Y6O
 
 | | Cryo-EM Structure of the human minor pre-B complex (pre-precatalytic spliceosome) U11 and tri-snRNP part | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Centrosomal AT-AC splicing factor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bai, R, Yuan, M, Zhang, P, Luo, T, Shi, Y, Wan, R. | | Deposit date: | 2024-02-02 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural basis of U12-type intron engagement by the fully assembled human minor spliceosome.
Science, 383, 2024
|
|
4E15
 
 | |
5F0A
 
 | | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine INHIBITOR | | Descriptor: | 1-tert-butyl-3-(3-chlorophenoxy)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, cGMP-dependent protein kinase, putative | | Authors: | Walker, J.R, Wernimont, A.K, He, H, Seitova, A, Loppnau, P, Sibley, L.D, Graslund, S, Hutchinson, A, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Hui, R, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CRYSTAL STRUCTURE OF PVX_084705 WITH BOUND INHIBITOR
To be published
|
|
5VS5
 
 | | ABA-mimicking ligand AMF2alpha in complex with ABA receptor PYL2 and PP2C HAB1 | | Descriptor: | 1-(2,3-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | Authors: | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-W, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.E, Zhu, J.-K. | | Deposit date: | 2017-05-11 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
