4DR2
 
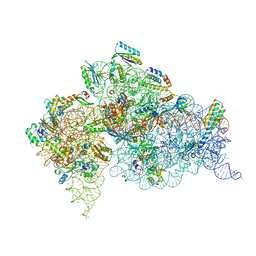 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
5I3G
 
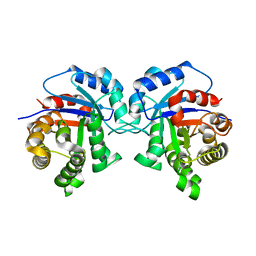 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
1T8W
 
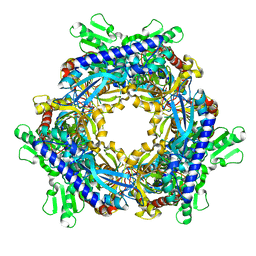 | |
1O9W
 
 | | F17-aG lectin domain from Escherichia coli in complex with N-acetyl-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17A-G FIMBRIAL ADHESIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1BRW
 
 | |
3IJL
 
 | | Structure of dipeptide epimerase from Bacteroides thetaiotaomicron complexed with L-Pro-D-Glu; nonproductive substrate binding. | | Descriptor: | D-GLUTAMIC ACID, MAGNESIUM ION, Muconate cycloisomerase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Lukk, T, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-08-04 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8PZ6
 
 | | crystal structure of VDR in complex with D-Bishomo-1a,25-dihydroxyvitamin D3 analog 56 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(4~{a}~{R},5~{S},9~{a}~{S})-4~{a}-methyl-5-[(2~{R})-6-methyl-6-oxidanyl-heptan-2-yl]-3,4,5,6,7,8,9,9~{a}-octahydro-2~{H}-benzo[7]annulen-1-ylidene]ethylidene]-2-(3-oxidanylpropylidene)cyclohexane-1,3-diol, Nuclear receptor coactivator 2, Vitamin D3 receptor A | | Authors: | Rochel, N. | | Deposit date: | 2023-07-27 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological activity of D-bishomo-1 alpha ,25-dihydroxyvitamin D 3 analogs and their crystal structures with the vitamin D nuclear receptor.
Eur.J.Med.Chem., 271, 2024
|
|
2VRH
 
 | | Structure of the E. coli trigger factor bound to a translating ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L23, 50S RIBOSOMAL PROTEIN L24, 50S RIBOSOMAL PROTEIN L29, ... | | Authors: | Merz, F, Boehringer, D, Schaffitzel, C, Preissler, S, Hoffmann, A, Maier, T, Rutkowska, A, Lozza, J, Ban, N, Bukau, B, Deuerling, E. | | Deposit date: | 2008-04-07 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Molecular Mechanism and Structure of Trigger Factor Bound to the Translating Ribosome.
Embo J., 27, 2008
|
|
2VVP
 
 | | Crystal structure of Mycobacterium tuberculosis ribose-5-phosphate isomerase B in complex with its substrates ribose 5-phosphate and ribulose 5-phosphate | | Descriptor: | 5-O-phosphono-D-ribose, RIBOSE-5-PHOSPHATE ISOMERASE B, RIBULOSE-5-PHOSPHATE | | Authors: | Kowalinski, E, Roos, A.K, Mariano, S, Salmon, L, Mowbray, S.L. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | D-Ribose-5-Phosphate Isomerase B from Escherichia Coli is Also a Functional D-Allose-6-Phosphate Isomerase, While the Mycobacterium Tuberculosis Enzyme is not.
J.Mol.Biol., 382, 2008
|
|
5J2U
 
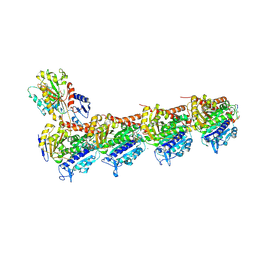 | | Tubulin-MMAF complex | | Descriptor: | CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Waight, A.B, Bargsten, K, Doronina, S, Steinmetz, M.O, Sussman, D, Prota, A.E. | | Deposit date: | 2016-03-30 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Microtubule Destabilization by Potent Auristatin Anti-Mitotics.
Plos One, 11, 2016
|
|
5J62
 
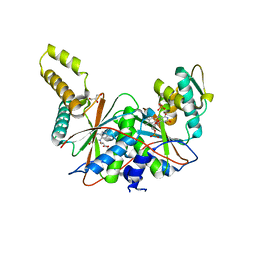 | | FMN-dependent Nitroreductase (CDR20291_0684) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Wang, B, Powell, S.M, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
6XAE
 
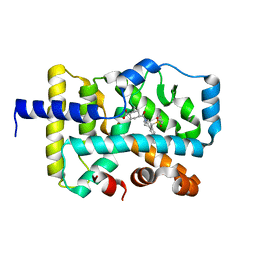 | | SUBSTITUTED BENZYLOXYTRICYCLIC COMPOUNDS AS RETINOIC ACID-RELATED ORPHAN RECEPTOR GAMMA T AGONISTS | | Descriptor: | Nuclear receptor ROR-gamma, trans-4-{(3aR,9bR)-7-[(2-chloro-6-fluorophenyl)methoxy]-9b-[(4-fluorophenyl)sulfonyl]-1,2,3a,4,5,9b-hexahydro-3H-benzo[e]indole-3-carbonyl}cyclohexane-1-carboxylic acid | | Authors: | Sack, J.S. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.257 Å) | | Cite: | Substituted benzyloxytricyclic compounds as retinoic acid-related orphan receptor gamma t (ROR gamma t) agonists.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4D8J
 
 | | Structure of E. coli MatP-mats complex | | Descriptor: | 5'-D(*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*A)-3', 5'-D(*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*A)-3', Macrodomain Ter protein | | Authors: | Dupaigne, P, Tonthat, N.K, Espeli, O, Whitfill, T, Boccard, F, Schumacher, M.A. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
1BJX
 
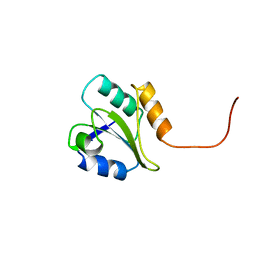 | | HUMAN PROTEIN DISULFIDE ISOMERASE, NMR, 24 STRUCTURES | | Descriptor: | PROTEIN DISULFIDE ISOMERASE | | Authors: | Kemmink, J, Dijkstra, K, Mariani, M, Scheek, R.M, Penka, E, Nilges, M, Darby, N.J. | | Deposit date: | 1998-06-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure in solution of the b domain of protein disulfide isomerase.
J.Biomol.NMR, 13, 1999
|
|
5JIK
 
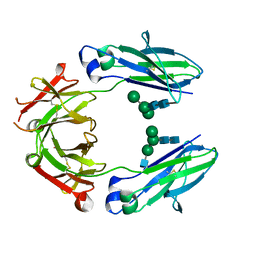 | | Crystal structure of HER2 binding IgG1-Fc (Fcab H10-03-6) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Humm, A, Lobner, E, Mlynek, G, Obinger, C, Djinovic-Carugo, K. | | Deposit date: | 2016-04-22 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fcab-HER2 Interaction: a Menage a Trois. Lessons from X-Ray and Solution Studies.
Structure, 25, 2017
|
|
4IM4
 
 | |
1AGC
 
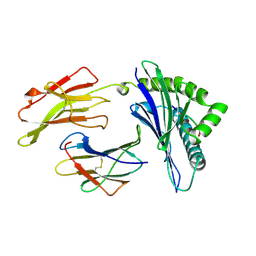 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
5IR4
 
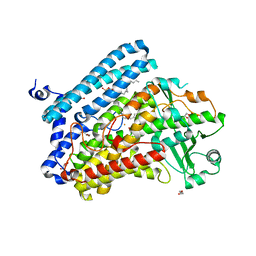 | | Crystal structure of wild-type bacterial lipoxygenase from Pseudomonas aeruginosa PA-LOX with space group C2221 at 1.48 A resolution | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradec-5-enoyloxy)propyl (11Z)-octadec-11-enoate, Arachidonate 15-lipoxygenase, CHLORIDE ION, ... | | Authors: | Kalms, J, Banthiya, S, Galemou Yoga, E, Kuhn, H, Scheerer, P. | | Deposit date: | 2016-03-12 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and functional basis of phospholipid oxygenase activity of bacterial lipoxygenase from Pseudomonas aeruginosa.
Biochim.Biophys.Acta, 1861, 2016
|
|
1O9Z
 
 | | F17-aG lectin domain from Escherichia coli (ligand free) | | Descriptor: | F17-AG LECTIN DOMAIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-23 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
5I3I
 
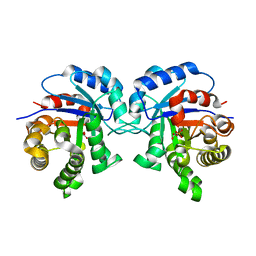 | | Structure-Function Studies on Role of Hydrophobic Clamping of a Basic Glutamate in Catalysis by Triosephosphate Isomerase | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase, glycosomal | | Authors: | Drake, E.J, Gulick, A.M, Richard, J.P, Zhai, X, Kim, K, Reinhardt, C.J. | | Deposit date: | 2016-02-10 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Function Studies of Hydrophobic Residues That Clamp a Basic Glutamate Side Chain during Catalysis by Triosephosphate Isomerase.
Biochemistry, 55, 2016
|
|
1YY4
 
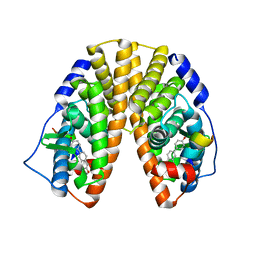 | | Crystal structure of estrogen receptor beta complexed with 1-chloro-6-(4-hydroxy-phenyl)-naphthalen-2-ol | | Descriptor: | 1-CHLORO-6-(4-HYDROXYPHENYL)-2-NAPHTHOL, Estrogen receptor beta, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Mewshaw, R.E, Edsall Jr, R.J, Yang, C, Manas, E.S, Xu, Z.B, Henderson, R.A, Keith Jr, J.C, Harris, H.A. | | Deposit date: | 2005-02-23 | | Release date: | 2006-02-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ERbeta ligands. 3. Exploiting two binding orientations of the 2-phenylnaphthalene scaffold to achieve ERbeta selectivity
J.Med.Chem., 48, 2005
|
|
1C6O
 
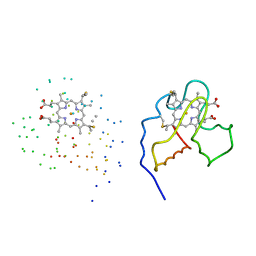 | | CRYSTAL STRUCTURE OF OXIDIZED CYTOCHROME C6 FROM THE GREEN ALGAE SCENEDESMUS OBLIQUUS | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schnackenberg, J, Than, M.E, Mann, K, Wiegand, G, Huber, R, Reuter, W. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino acid sequence, crystallization and structure determination of reduced and oxidized cytochrome c6 from the green alga Scenedesmus obliquus.
J.Mol.Biol., 290, 1999
|
|
2X97
 
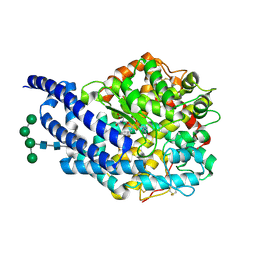 | | Crystal structure of AnCE-RXP407 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANGIOTENSIN CONVERTING ENZYME, N~2~-acetyl-N-{(1R)-1-[(S)-[(2S)-3-{[(2S)-1-amino-1-oxopropan-2-yl]amino}-2-methyl-3-oxopropyl](hydroxy)phosphoryl]-2-phenylethyl}-L-alpha-asparagine, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
2X92
 
 | | Crystal structure of AnCE-ramiprilat complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ANGIOTENSIN CONVERTING ENZYME, ... | | Authors: | Akif, M, Georgiadis, D, Mahajan, A, Dive, V, Sturrock, E.D, Isaac, R.E, Acharya, K.R. | | Deposit date: | 2010-03-14 | | Release date: | 2010-06-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | High Resolution Crystal Structures of Drosophila Melanogaster Angiotensin Converting Enzyme in Complex with Novel Inhibitors and Anti- Hypertensive Drugs.
J.Mol.Biol., 400, 2010
|
|
4DR4
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, cognate transfer RNA anticodon stem-loop and multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.969 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
