4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4DK7
 
 | | Crystal structure of LXR ligand binding domain in complex with full agonist 1 | | Descriptor: | ACETATE ION, CALCIUM ION, N-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]-N-methylbenzenesulfonamide, ... | | Authors: | Piper, D.E, Xu, H. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2012-05-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of a new binding mode for a series of liver X receptor agonists.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6YND
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 1 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.525 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6YNF
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 3 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
4DOW
 
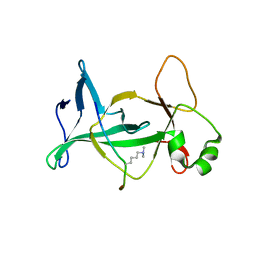 | | Structure of mouse ORC1 BAH domain bound to H4K20me2 | | Descriptor: | Histone H4, Origin recognition complex subunit 1 | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2012-02-10 | | Release date: | 2012-03-07 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome.
Nature, 484, 2012
|
|
3LBD
 
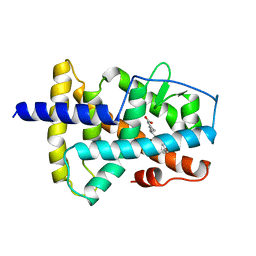 | | LIGAND-BINDING DOMAIN OF THE HUMAN RETINOIC ACID RECEPTOR GAMMA BOUND TO 9-CIS RETINOIC ACID | | Descriptor: | (9cis)-retinoic acid, RETINOIC ACID RECEPTOR GAMMA | | Authors: | Klaholz, B.P, Renaud, J.-P, Mitschler, A, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 1998-02-04 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational adaptation of agonists to the human nuclear receptor RAR gamma.
Nat.Struct.Biol., 5, 1998
|
|
3LRE
 
 | |
4DGC
 
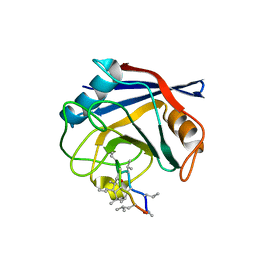 | | TRIMCyp cyclophilin domain from Macaca mulatta: cyclosporin A complex | | Descriptor: | TRIMCyp, cyclosporin A | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6YGJ
 
 | |
4Z97
 
 | |
6Y9Z
 
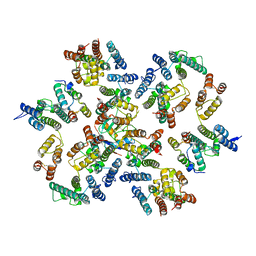 | | Structure of the native full-length HIV-1 capsid protein in complex with Cyclophilin A from helical assembly (-13,9) | | Descriptor: | Gag-Pol polyprotein, Peptidyl-prolyl cis-trans isomerase A | | Authors: | Ni, T, Gerard, S, Zhao, G, Ning, J, Zhang, P. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-19 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
4NH1
 
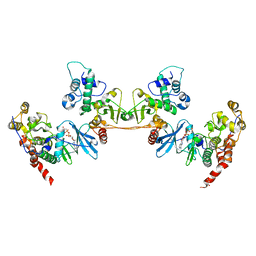 | | Crystal structure of a heterotetrameric CK2 holoenzyme complex carrying the Andante-mutation in CK2beta and consistent with proposed models of autoinhibition and trans-autophosphorylation | | Descriptor: | Casein kinase II subunit alpha, Casein kinase II subunit beta, GLYCEROL, ... | | Authors: | Schnitzler, A, Issinger, O.-G, Niefind, K. | | Deposit date: | 2013-11-04 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The Protein Kinase CK2(Andante) Holoenzyme Structure Supports Proposed Models of Autoregulation and Trans-Autophosphorylation
J.Mol.Biol., 426, 2014
|
|
4DGD
 
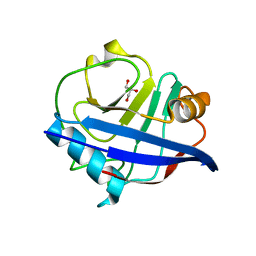 | | TRIMCyp cyclophilin domain from Macaca mulatta: H70C mutant | | Descriptor: | GLYCEROL, TRIMCyp | | Authors: | Caines, M.E.C, Bichel, K, Price, A.J, McEwan, W.A, James, L.C. | | Deposit date: | 2012-01-25 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse HIV viruses are targeted by a conformationally dynamic antiviral.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6YBV
 
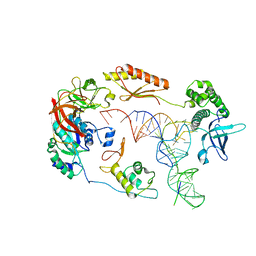 | | Structure of a human 48S translational initiation complex - eIF2-TC | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
4DGL
 
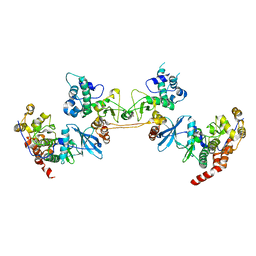 | | Crystal Structure of the CK2 Tetrameric Holoenzyme | | Descriptor: | Casein kinase II subunit alpha, Casein kinase II subunit beta, ZINC ION | | Authors: | Lolli, G, Battistutta, R. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural determinants of protein kinase CK2 regulation by autoinhibitory polymerization.
Acs Chem.Biol., 7, 2012
|
|
4X2I
 
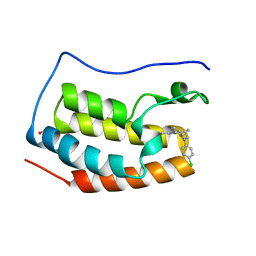 | | Discovery of benzotriazolo diazepines as orally-active inhibitors of BET bromodomains: Crystal structure of BRD4 with CPI-13 | | Descriptor: | (4R)-6-(4-chlorophenyl)-1,4-dimethyl-5,6-dihydro-4H-[1,2,4]triazolo[4,3-a][1,5]benzodiazepine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Bellon, S.F, Jayaram, H, Poy, F. | | Deposit date: | 2014-11-26 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Discovery of Benzotriazolo[4,3-d][1,4]diazepines as Orally Active Inhibitors of BET Bromodomains.
Acs Med.Chem.Lett., 7, 2016
|
|
3UAL
 
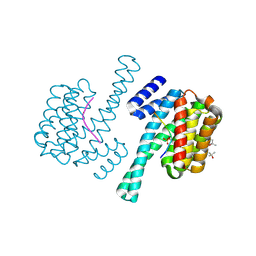 | |
3KFC
 
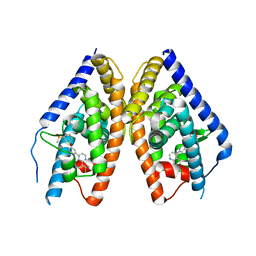 | | Complex Structure of LXR with an agonist | | Descriptor: | 4-{3-[3-(methylsulfonyl)phenoxy]phenyl}-8-(trifluoromethyl)quinoline, Oxysterols receptor LXR-beta | | Authors: | Olland, A, Bernotas, R.C, Unwalla, R. | | Deposit date: | 2009-10-27 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 4-(3-Aryloxyaryl)quinoline sulfones are potent liver X receptor agonists.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3R21
 
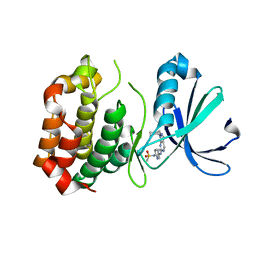 | | Design, synthesis, and biological evaluation of pyrazolopyridine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (Part I) | | Descriptor: | MAGNESIUM ION, N-(2-aminoethyl)-N-{5-[(1-cycloheptyl-1H-pyrazolo[3,4-d]pyrimidin-6-yl)amino]pyridin-2-yl}methanesulfonamide, Serine/threonine-protein kinase 6 | | Authors: | Zhang, L, Fan, J, Chong, J.-H, Cesena, A, Tam, B, Gilson, C, Boykin, C, Wang, D, Marcotte, D, Le Brazidec, J.-Y, Aivazian, D, Piao, J, Lundgren, K, Hong, K, Vu, K, Nguyen, K. | | Deposit date: | 2011-03-11 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Design, synthesis, and biological evaluation of pyrazolopyrimidine-sulfonamides as potent multiple-mitotic kinase (MMK) inhibitors (part I).
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4MEN
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 5-methyl-triazolopyrimidine ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,5-dimethyl-N-(4-methylbenzyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
4MI5
 
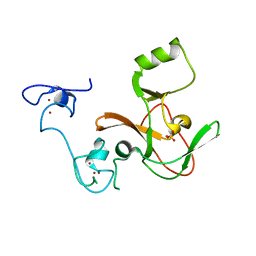 | | Crystal structure of the EZH2 SET domain | | Descriptor: | Histone-lysine N-methyltransferase EZH2, SULFATE ION, ZINC ION | | Authors: | Antonysamy, S, Condon, B, Druzina, Z, Bonanno, J, Gheyi, T, Macewan, I, Zhang, A, Ashok, S, Russell, M, Luz, J.G. | | Deposit date: | 2013-08-30 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Context of Disease-Associated Mutations and Putative Mechanism of Autoinhibition Revealed by X-Ray Crystallographic Analysis of the EZH2-SET Domain.
Plos One, 8, 2013
|
|
4Z49
 
 | |
4Z7B
 
 | | Structure of the enzyme-product complex resulting from TDG action on a GfC mismatch | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DNA (28-MER), ... | | Authors: | Pozharski, E, Malik, S.S, Drohat, A.C. | | Deposit date: | 2015-04-07 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Thymine DNA glycosylase exhibits negligible affinity for nucleobases that it removes from DNA.
Nucleic Acids Res., 43, 2015
|
|
3RLO
 
 | | Structural Basis of Cytosolic DNA Recognition by Innate Receptors | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Gamma-interferon-inducible protein 16 | | Authors: | Jin, T.C, Xiao, T. | | Deposit date: | 2011-04-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the HIN Domain:DNA Complexes Reveal Ligand Binding and Activation Mechanisms of the AIM2 Inflammasome and IFI16 Receptor.
Immunity, 36, 2012
|
|
1SVC
 
 | | NFKB P50 HOMODIMER BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*GP*AP*TP*GP*GP*GP*GP*AP*AP*TP*CP*CP*CP*CP*TP*A P*GP*A)-3'), PROTEIN (NUCLEAR FACTOR KAPPA-B (NF-KB)) | | Authors: | Mueller, C.W, Harrison, S.C. | | Deposit date: | 1995-11-27 | | Release date: | 1996-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the NF-kappa B p50 homodimer bound to DNA.
Nature, 373, 1995
|
|
