6C0K
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.958 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
4S34
 
 | | ERK2 (I84A) in complex with AMP-PNP | | Descriptor: | MAGNESIUM ION, Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Livnah, O, Karamansha, Y, Tzarum, N. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Multiple mechanisms render Erk proteins MEK-independent
To be Published
|
|
3PDN
 
 | | Crystal structure of SmyD3 in complex with methyltransferase inhibitor sinefungin | | Descriptor: | GLYCEROL, MAGNESIUM ION, SET and MYND domain-containing protein 3, ... | | Authors: | Sirinupong, N, Brunzelle, J, Yang, Z. | | Deposit date: | 2010-10-23 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights into the Autoinhibition and Posttranslational Activation of Histone Methyltransferase SmyD3.
J.Mol.Biol., 406, 2011
|
|
6FTG
 
 | |
3K0E
 
 | | Crystal structure of the phosphorylation-site mutant T426N of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
6C0L
 
 | | Crystal structure of HIV-1 E138K mutant reverse transcriptase in complex with non-nucleoside inhibitor K-5a2 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)thieno[3,2-d]pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzene-1-sulfonamide, MAGNESIUM ION, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
4S32
 
 | |
8FC3
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and telithromycin at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
6FTJ
 
 | | Cryo-EM Structure of the Mammalian Oligosaccharyltransferase Bound to Sec61 and the Non-programmed 80S Ribosome | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Braunger, K, Becker, T, Beckmann, R. | | Deposit date: | 2018-02-22 | | Release date: | 2018-03-21 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum.
Science, 360, 2018
|
|
8EV6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with amikacin, mRNA, and A-, P-, and E-site tRNAs | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Seely, S.M, Gagnon, M.G. | | Deposit date: | 2022-10-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.946 Å) | | Cite: | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome.
Nat Commun, 14, 2023
|
|
8EV7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with kanamycin, mRNA, and A-, P-, and E-site tRNAs | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seely, S.M, Gagnon, M.G. | | Deposit date: | 2022-10-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome.
Nat Commun, 14, 2023
|
|
8FC2
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
5UMO
 
 | |
8FC5
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and azithromycin at 2.65A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
8FC4
 
 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with protein Y, hygromycin A, and erythromycin at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Chen, C.-W, Syroegin, E.A, Svetlov, M.S, Polikanov, Y.S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural insights into the mechanism of overcoming Erm-mediated resistance by macrolides acting together with hygromycin-A.
Nat Commun, 14, 2023
|
|
2CHG
 
 | | Replication Factor C domains 1 and 2 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | Authors: | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
3RJR
 
 | | Crystal Structure of pro-TGF beta 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Transforming growth factor beta-1 | | Authors: | Zhu, J.H, Shi, M.L, Springer, T.A. | | Deposit date: | 2011-04-15 | | Release date: | 2011-06-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Latent TGF-Beta structure and activation
Nature, 474, 2011
|
|
3JZM
 
 | | Crystal structure of the phosphorylation-site mutant T432A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase kaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-23 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
2ZB1
 
 | | Crystal structure of P38 in complex with biphenyl amide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(cyclopropylmethyl)-2'-methyl-5'-(5-methyl-1,3,4-oxadiazol-2-yl)biphenyl-4-carboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2007-10-13 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 1: Discovery and binding mode
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4S30
 
 | |
6XXU
 
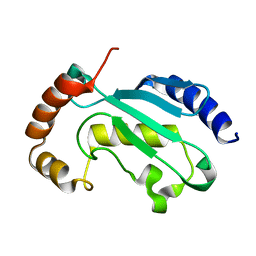 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
8GJY
 
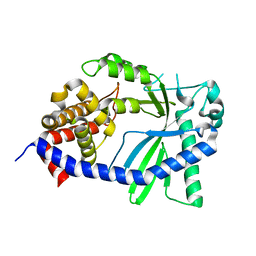 | | Structure of a cGAS-like receptor Sp-cGLR1 from S. pistillata | | Descriptor: | cGAS-like receptor 1 | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJW
 
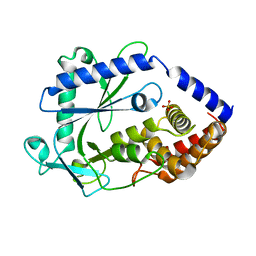 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
4RUO
 
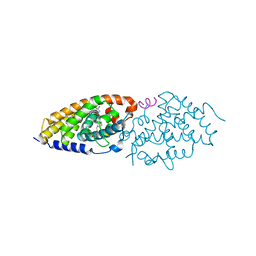 | | Crystal structure of zVDR L337H mutant-gemini complex | | Descriptor: | 21-NOR-9,10-SECOCHOLESTA-5,7,10(19)-TRIENE-1,3,25-TRIOL, 20-(4-HYDROXY-4-METHYLPENTYL)-, (1A,3B,5Z,7E), ... | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
8GJX
 
 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
