7Z3N
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-1 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3O
 
 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-2 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ZW0
 
 | | FAP-80S Complex - Rotated state | | Descriptor: | 18S ribosomal RNA (RDN18-1), 25S ribosomal RNA (RDN25-1), 40S ribosomal protein S0-A, ... | | Authors: | Ikeuchi, K, Buschauer, R, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Sensing of individual stalled 80S ribosomes by Fap1 for nonfunctional rRNA turnover.
Mol.Cell, 82, 2022
|
|
7ZQ9
 
 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.74 A resolution (symmetry expanded) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQC
 
 | | Monomeric PSI of Chlamydomonas reinhardtii at 2.31 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZQD
 
 | | Dimeric PSI of Chlamydomonas reinhardtii at 2.97 A resolution | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Naschberger, A, Amunts, A. | | Deposit date: | 2022-04-29 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Algal photosystem I dimer and high-resolution model of PSI-plastocyanin complex.
Nat.Plants, 8, 2022
|
|
7ZTA
 
 | | Structure of an Escherichia coli 70S ribosome stalled by Tetracenomycin X during translation of an MAAAPQK(C) peptide | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Leroy, E.C, Perry, T.N, Renault, T.T, Innis, C.A. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Tetracenomycin X sequesters peptidyl-tRNA during translation of QK motifs.
Nat.Chem.Biol., 19, 2023
|
|
8A3L
 
 | | Structural insights into the binding of bS1 to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | D'Urso, G, Chat, S, Gillet, R, Giudice, E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the binding of bS1 to the ribosome.
Nucleic Acids Res., 51, 2023
|
|
8B0X
 
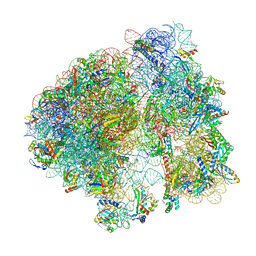 | | Translating 70S ribosome in the unrotated state (P and E, tRNAs) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fromm, S.A, O'Connor, K.M, Purdy, M, Bhatt, P.R, Loughran, G, Atkins, J.F, Jomaa, A, Mattei, S. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-30 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | The translating bacterial ribosome at 1.55 angstrom resolution generated by cryo-EM imaging services.
Nat Commun, 14, 2023
|
|
8A3D
 
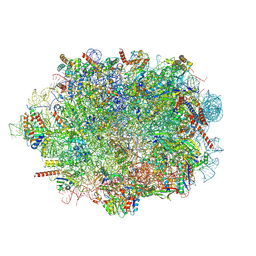 | | Human mature large subunit of the ribosome with eIF6 and homoharringtonine bound | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 28S ribosomal RNA, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Faille, A, Warren, A.J, Dent, K.C. | | Deposit date: | 2022-06-08 | | Release date: | 2023-03-08 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | The chemical landscape of the human ribosome at 1.67 angstrom resolution.
Biorxiv, 2023
|
|
8AKN
 
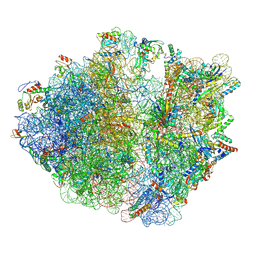 | | Cryo-EM structure of the proline-rich antimicrobial peptide drosocin bound to the terminating ribosome | | Descriptor: | 16S ribosomal RNA, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 23S ribosomal RNA, ... | | Authors: | Koller, T.O, Morici, M, Wilson, D.N. | | Deposit date: | 2022-07-30 | | Release date: | 2023-03-08 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for translation inhibition by the glycosylated drosocin peptide.
Nat.Chem.Biol., 19, 2023
|
|
8B2L
 
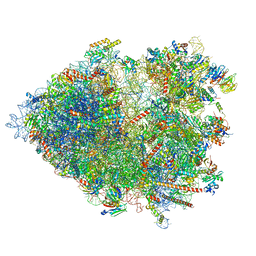 | | Cryo-EM structure of the plant 80S ribosome | | Descriptor: | 18S rRNA, 25S rRNA, 30S ribosomal protein S15, ... | | Authors: | Smirnova, J, Loerke, J, Kleinau, G, Schmidt, A, Buerger, J, Meyer, E.H, Mielke, T, Scheerer, P, Bock, R, Spahn, C.M.T, Zoschke, R. | | Deposit date: | 2022-09-14 | | Release date: | 2023-08-23 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structure of the actively translating plant 80S ribosome at 2.2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8ANY
 
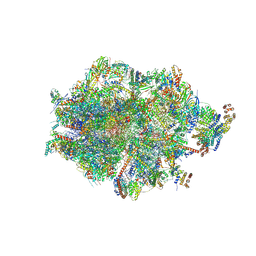 | | Human mitochondrial ribosome in complex with LRPPRC, SLIRP, A-site, P-site, E-site tRNAs and mRNA | | Descriptor: | 1,4-DIAMINOBUTANE, 12S mitochondrial rRNA, 16S mitochondrial rRNA, ... | | Authors: | Singh, V, Itoh, Y, Amunts, A. | | Deposit date: | 2022-08-06 | | Release date: | 2023-08-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of LRPPRC-SLIRP-dependent translation by the
mitoribosome
To Be Published
|
|
2XO7
 
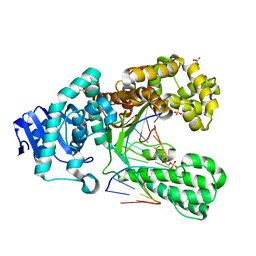 | | Crystal structure of a dA:O-allylhydroxylamine-dC basepair in complex with fragment DNA polymerase I from Bacillus stearothermophilus | | Descriptor: | 5'-D(*AP*GP*GP*AP*AP*TP*GP*GP*TP*CP*A)-3', 5'-D(*GP*AP*CP*CP*AP*TP*47C*CP*CP*T)-3', DNA POLYMERASE I, ... | | Authors: | Muenzel, M, Lercher, L, Mueller, M, Carell, T. | | Deposit date: | 2010-08-10 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Chemical Discrimination between Dc and 5Medc Via Their Hydroxylamine Adducts
Nucleic Acids Res., 38, 2010
|
|
2Y5Z
 
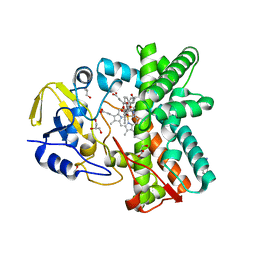 | | Mixed-function P450 MycG in complex with mycinamicin III in C2221 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN III, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-19 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y46
 
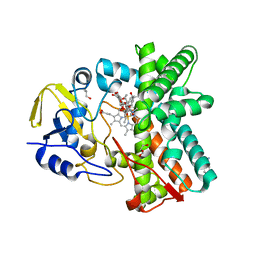 | | Structure of the mixed-function P450 MycG in complex with mycinamicin IV in C 2 2 21 space group | | Descriptor: | BENZAMIDINE, GLYCEROL, MYCINAMICIN IV, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
2Y5N
 
 | | Structure of the mixed-function P450 MycG in complex with mycinamicin V in P21 space group | | Descriptor: | GLYCEROL, MAGNESIUM ION, MYCINAMICIN V, ... | | Authors: | Li, S, Kells, P.M, Sherman, D.H, Podust, L.M. | | Deposit date: | 2011-01-15 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Substrate Recognition by the Multifunctional Cytochrome P450 Mycg in Mycinamicin Hydroxylation and Epoxidation Reactions.
J.Biol.Chem., 287, 2012
|
|
8DYB
 
 | |
4OQS
 
 | | Crystal structure of CYP105AS1 | | Descriptor: | CYP105AS1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Leys, D. | | Deposit date: | 2014-02-10 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8E5J
 
 | | The crystal structure of 4-n-butylbenzoic acid bound CYP199A4 | | Descriptor: | 4-butylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Doherty, D.Z, Bell, S.G, Bruning, J.B. | | Deposit date: | 2022-08-22 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Exploring the Factors which Result in Cytochrome P450 Catalyzed Desaturation Versus Hydroxylation.
Chem Asian J, 17, 2022
|
|
6DAQ
 
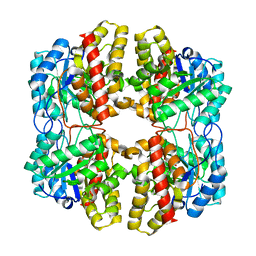 | | PhdJ bound to substrate intermediate | | Descriptor: | PhdJ | | Authors: | Medellin, B.P, LeVieux, J.A, Zhang, Y.J, Whitman, C.P. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Hydratase-Aldolases, NahE and PhdJ: Implications for the Specificity, Catalysis, and N-Acetylneuraminate Lyase Subgroup of the Aldolase Superfamily.
Biochemistry, 57, 2018
|
|
6DHX
 
 | |
7LTN
 
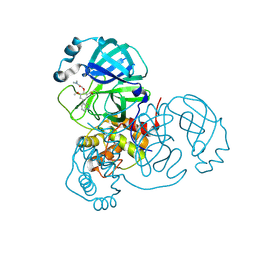 | | Crystal structure of Mpro in complex with inhibitor CDD-1713 | | Descriptor: | 2-[4-(1~{H}-indazol-4-yl)-2-methanoyl-6-methoxy-phenoxy]-~{N},~{N}-dimethyl-ethanamide, 3C-like proteinase | | Authors: | Lu, S, Palzkill, T, Matzuk, M, Young, D, Melek, N, Chamakuri, S. | | Deposit date: | 2021-02-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 M pro inhibitors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5D0B
 
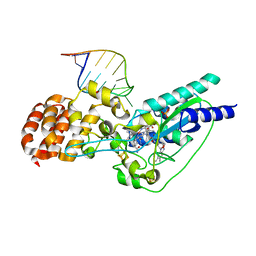 | | Crystal structure of epoxyqueuosine reductase with a tRNA-TYR epoxyqueuosine-modified tRNA stem loop | | Descriptor: | COBALAMIN, Epoxyqueuosine reductase, GLYCEROL, ... | | Authors: | Dowling, D.P, Miles, Z.D, Kohrer, C, Bandarian, V, Drennan, C.L. | | Deposit date: | 2015-08-03 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Molecular basis of cobalamin-dependent RNA modification.
Nucleic Acids Res., 44, 2016
|
|
5CX1
 
 | | Nitrogenase molybdenum-iron protein beta-K400E mutant | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Owens, C.P, Luca, M.A, Tezcan, F.A. | | Deposit date: | 2015-07-28 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7476 Å) | | Cite: | Evidence for Functionally Relevant Encounter Complexes in Nitrogenase Catalysis.
J.Am.Chem.Soc., 137, 2015
|
|
