7N47
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7ZQR
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 4-(4-methoxyphenyl)pyridine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snee, M, Katariya, M, Tunnicliffe, R, Levy, C, Leys, D. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
6JVC
 
 | | Structure of the Cobalt Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-04-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7N55
 
 | | The crystal structure of the mutant I38T PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988514 | | Descriptor: | 5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6C1C
 
 | | FGFR1 kinase complex with inhibitor SN37116 | | Descriptor: | 7-(cyclohexylamino)-3-(2,6-dichloro-3,5-dimethoxyphenyl)-1-{(3S)-1-[(2E)-4-(dimethylamino)but-2-enoyl]pyrrolidin-3-yl}-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one, Fibroblast growth factor receptor 1, SULFATE ION | | Authors: | Yosaatmadja, Y, Smaill, J.B, Squire, C.J. | | Deposit date: | 2018-01-04 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Understanding the structural requirements for covalent inhibition of FGFR1-3
To Be Published
|
|
6C1J
 
 | | Crystal Structure of Ketosteroid Isomerase Y32F/Y57F/D40N mutant from Pseudomonas Putida (pKSI) bound to 3,4-dinitrophenol | | Descriptor: | 3,4-dinitrophenol, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Yabukarski, F, Pinney, M.M, Herschlag, D. | | Deposit date: | 2018-01-04 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.063 Å) | | Cite: | Structural Coupling Throughout the Active Site Hydrogen Bond Networks of Ketosteroid Isomerase and Photoactive Yellow Protein.
J. Am. Chem. Soc., 140, 2018
|
|
7ZSU
 
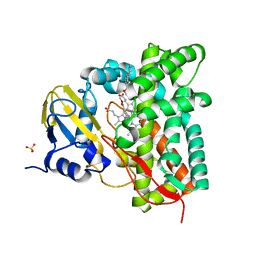 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, Steroid C26-monooxygenase, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-09 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
8T8A
 
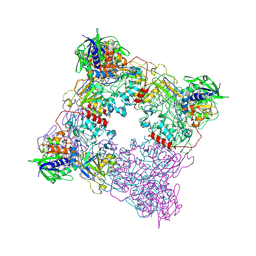 | |
7N4F
 
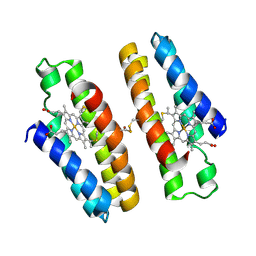 | |
7BJ4
 
 | | Inulosucrase from Halalkalicoccus jeotgali bound to kestose | | Descriptor: | Levansucrase, beta-D-fructofuranose-(2-1)-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
7N4G
 
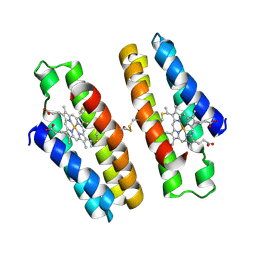 | |
7BJ5
 
 | | Inulosucrase from Halalkalicoccus jeotgali | | Descriptor: | Levansucrase | | Authors: | Ghauri, K, Pijning, T, Munawar, N, Ali, H, Ghauri, M.A, Anwar, M.A, Wallis, R. | | Deposit date: | 2021-01-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of an inulosucrase from Halalkalicoccus jeotgali B3T, a halophilic archaeal strain.
Febs J., 288, 2021
|
|
7ZXD
 
 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with an inhibitor | | Descriptor: | 1-[1-(2-piperidin-4-ylethyl)-5-pyridin-4-yl-indol-2-yl]butan-1-one, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Snee, M, Katariya, M, Levy, C, Leys, D. | | Deposit date: | 2022-05-20 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7N68
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988288 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7BIR
 
 | | Inhibitor of MDM2-p53 Interaction | | Descriptor: | 1-[[(1~{R})-2-[(5-chloranylpyridin-2-yl)methyl]-1-(4-chlorophenyl)-7-fluoranyl-3-oxidanylidene-5-(2-oxidanylpropan-2-yl)isoindol-1-yl]oxymethyl]cyclopropane-1-carboxamide, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Williams, P.A. | | Deposit date: | 2021-01-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-Based Design of Potent and Orally Active Isoindolinone Inhibitors of MDM2-p53 Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
8TDE
 
 | |
7ZW3
 
 | |
8TYX
 
 | |
6C2D
 
 | |
7BUG
 
 | | Reduced oxygenase of carbazole 1,9a-dioxygenase | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, CARBONATE ION, ... | | Authors: | Matsuzawa, J, Wang, Y.X, Suzuki-Minakuchi, C, Nojiri, H. | | Deposit date: | 2020-04-06 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Reduced oxygenase of carbazole 1,9a-dioxygenase
To Be Published
|
|
8A4E
 
 | |
8A2W
 
 | |
6JXM
 
 | | Crystal Structure of phi29 pRNA domain II | | Descriptor: | BARIUM ION, MAGNESIUM ION, RNA (97-mer) | | Authors: | Cai, R, Price, I.R, Ding, F, Wu, F, Chen, T, Zhang, Y, Liu, G, Jardine, P.J, Lu, C, Ke, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor.
Nucleic Acids Res., 47, 2019
|
|
7BVR
 
 | | DgpB-DgpC complex apo | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AP_endonuc_2 domain-containing protein, DgpB, ... | | Authors: | Mori, T, He, H, Abe, I. | | Deposit date: | 2020-04-11 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | C-Glycoside metabolism in the gut and in nature: Identification, characterization, structural analyses and distribution of C-C bond-cleaving enzymes.
Nat Commun, 12, 2021
|
|
7MVX
 
 | | Crystal structure of the Chaetomium thermophilum Nup188-Nic96 complex (Nup188 residues 1-1858; Nic96 residues 240-301) | | Descriptor: | Nucleoporin NIC96, Nucleoporin NUP188 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
