8RG9
 
 | | High pH (8.0) nitrite-bound MSOX movie series dataset 2 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [1.36 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, GLYCEROL, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
6GG8
 
 | | Mineralocorticoid receptor in complex with (s)-13 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Mineralocorticoid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Edman, K, Aagaard, A, Tangefjord, S. | | Deposit date: | 2018-05-03 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Mineralocorticoid Receptor Modulators with Low Impact on Electrolyte Homeostasis but Maintained Organ Protection.
J.Med.Chem., 62, 2019
|
|
7G0F
 
 | | Crystal Structure of human FABP4 in complex with 6-[(3-methoxycarbonyl-4-thiophen-2-ylthiophen-2-yl)carbamoyl]cyclohex-3-ene-1-carboxylic acid, i.e. SMILES C1(=C(NC(=O)[C@H]2[C@@H](C(=O)O)CC=CC2)SC=C1C1=CC=CS1)C(=O)OC with IC50=1.1 microM | | Descriptor: | (1R,2S)-2-{[(2M)-4'-(methoxycarbonyl)-4,5-dihydro-2'H,3H-[2,3'-bi-1lambda~4~-thiophen]-5'-yl]carbamoyl}cyclohexane-1-carboxylic acid, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3JTX
 
 | |
6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
7Y7T
 
 | | QDE-1 in complex with 12nt DNA template, ATP and 3'-dGTP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*GP*AP*G*AP*GP*AP*CP*CP*TP*TP*TP*T)-3'), GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Cui, R.X, Gan, J.H, Ma, J.B. | | Deposit date: | 2022-06-22 | | Release date: | 2022-09-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the dual activities of the two-barrel RNA polymerase QDE-1.
Nucleic Acids Res., 50, 2022
|
|
5ENC
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with N-(2,6-Dichlorobenzyl)acetamide (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, PH-interacting protein, ~{N}-[[2,6-bis(chloranyl)phenyl]methyl]ethanamide | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Szykowska, A, Burgess-Brown, N, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
150D
 
 | | GUANINE.1,N6-ETHENOADENINE BASE-PAIRS IN THE CRYSTAL STRUCTURE OF D(CGCGAATT(EDA)GCG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(EDA)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Leonard, G.A, McAuley-Hecht, K.E, Gibson, N.J, Brown, T, Watson, W.P, Hunter, W.N. | | Deposit date: | 1993-12-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine-1,N6-ethenoadenine base pairs in the crystal structure of d(CGCGAATT(epsilon dA)GCG).
Biochemistry, 33, 1994
|
|
6H06
 
 | | FAB CBTAU-22.1 IN COMPLEX WITH TAU PEPTIDE V1088-5 | | Descriptor: | GLYCEROL, HUMAN FAB ANTIBODY FRAGMENT OF CBTAU-22.1, HUMAN FAB ANTIBODY FRAGMENT OF HCBTAU-22.1, ... | | Authors: | Juraszek, J, Steinbacher, S. | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Enhancement of therapeutic potential of a naturally occurring human antibody targeting a phosphorylated Ser422containing epitope on pathological tau.
Acta Neuropathol Commun, 6, 2018
|
|
1JV6
 
 | | BACTERIORHODOPSIN D85S/F219L DOUBLE MUTANT AT 2.00 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
7T88
 
 | | Crystal Structure of the C-terminal Domain of the Phosphate Acetyltransferase from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Kim, Y, Dementiev, A, Welk, L, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-12-15 | | Release date: | 2021-12-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of c from Escherichia coli
To Be Published
|
|
6H78
 
 | | E1 enzyme for ubiquitin like protein activation. | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Soudah, N, Padala, P, Hassouna, F, Mashahreh, B, Lebedev, A.A, Isupov, M.N, Cohen-Kfir, E, Wiener, R. | | Deposit date: | 2018-07-30 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An N-Terminal Extension to UBA5 Adenylation Domain Boosts UFM1 Activation: Isoform-Specific Differences in Ubiquitin-like Protein Activation.
J.Mol.Biol., 431, 2019
|
|
9BFY
 
 | | Tri-complex of Compound-14, KRAS G12C, and CypA | | Descriptor: | (3R)-N-[(2S)-1-{[(1M,8R,10R,14S,21M)-22-ethyl-4-hydroxy-21-{2-[(1R)-1-methoxyethyl]pyridin-3-yl}-18,18-dimethyl-9,15-dioxo-16-oxa-10,22,28-triazapentacyclo[18.5.2.1~2,6~.1~10,14~.0~23,27~]nonacosa-1(25),2(29),3,5,20,23,26-heptaen-8-yl]amino}-3-methyl-1-oxobutan-2-yl]-N-methyl-1-propanoylpyrrolidine-3-carboxamide (non-preferred name), CHLORIDE ION, GTPase KRas, ... | | Authors: | Tomlinson, A.C.A, Saldajeno-Concar, M, Knox, J.E, Yano, J.K. | | Deposit date: | 2024-04-18 | | Release date: | 2024-06-12 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of Elironrasib (RMC-6291), a Potent and Orally Bioavailable, RAS(ON) G12C-Selective, Covalent Tricomplex Inhibitor for the Treatment of Patients with RAS G12C-Addicted Cancers.
J.Med.Chem., 68, 2025
|
|
5AP3
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 9-CYCLOPENTYL-2-[[2-METHOXY-4-[(1-METHYLPIPERIDIN-4-YL)OXY]-PHENYL]AMINO]-7-METHYL-7,9-DIHYDRO-8H-PURIN-8-ONE, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
6XKG
 
 | | Crystal structure of 3-O-Sulfotransferase isoform 3 in complex with 8mer oligosaccharide with 6S sulfation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ... | | Authors: | Pedersen, L.C, Liu, J, Wander, R. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the substrate recognition mechanisms of the heparan sulfate 3- O -sulfotransferase-3.
Rsc Chem Biol, 2, 2021
|
|
4M1R
 
 | | Structure of a novel cellulase 5 from a sugarcane soil metagenomic library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellulase 5 | | Authors: | Paiva, J.H, Alvarez, T.M, Cairo, J.P, Paixao, D.A, Almeida, R.A, Tonoli, C.C.C, Ruiz, D.M, Ruller, R, Santos, C.R, Squina, F.M, Murakami, M.T. | | Deposit date: | 2013-08-03 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of a novel cellulase 5 from sugarcane soil metagenome.
Plos One, 8, 2013
|
|
6I6L
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydrocolumbamine | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
7VJL
 
 | | The crystal structure of FGFR4 kinase domain in complex with N-(5-cyano-4-((2-methoxyethyl)amino)pyridin-2-yl)-7-(2,2,2-trifluoroacetyl)-3,4-dihydro-1,8-naphthyridine-1(2H)-carboxamide | | Descriptor: | Fibroblast growth factor receptor 4, N-[5-(aminomethyl)-4-(2-methoxyethylamino)pyridin-2-yl]-7-[2,2,2-tris(fluoranyl)ethanoyl]-3,4-dihydro-2H-1,8-naphthyridine-1-carboxamide | | Authors: | Zhang, Z.M, Wang, Y.J. | | Deposit date: | 2021-09-28 | | Release date: | 2022-02-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.900173 Å) | | Cite: | Characterization of an aromatic trifluoromethyl ketone as a new warhead for covalently reversible kinase inhibitor design.
Bioorg.Med.Chem., 50, 2021
|
|
8T6F
 
 | | Crystal structure of human MBP-Myeloid cell leukemia 1 (Mcl-1) in complex with BRD810 inhibitor | | Descriptor: | (3aM,9S,15R)-4-chloro-3-ethyl-7-{3-[(6-fluoronaphthalen-1-yl)oxy]propyl}-2-methyl-15-[2-(morpholin-4-yl)ethyl]-2,10,11,12,13,15-hexahydropyrazolo[4',3':9,10][1,6]oxazacycloundecino[8,7,6-hi]indole-8-carboxylic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Poncet-Montange, G, Lemke, C.T. | | Deposit date: | 2023-06-15 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | BRD-810 is a highly selective MCL1 inhibitor with optimized in vivo clearance and robust efficacy in solid and hematological tumor models.
Nat Cancer, 5, 2024
|
|
8OGF
 
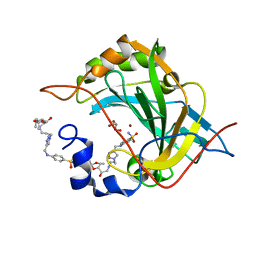 | | Human Carbonic Anhydrase II in complex with 4-(((1-(3-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)propyl)-1H-1,2,3-triazol-4-yl)methyl)amino)benzenesulfonamide | | Descriptor: | 4-[[1-[3-[[(2~{R})-2-oxidanyl-2-[(4~{S},5~{R})-2,2,5-trimethyl-1,3-dioxolan-4-yl]ethyl]amino]propyl]-1,2,3-triazol-4-yl]methylamino]benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Last modified: | 2025-10-08 | | Method: | X-RAY DIFFRACTION (1.321 Å) | | Cite: | Mono- and three-tailed sugar and iminosugar decorated benzenesulfonamide carbonic anhydrase inhibitors.
Org.Biomol.Chem., 21, 2023
|
|
8C3P
 
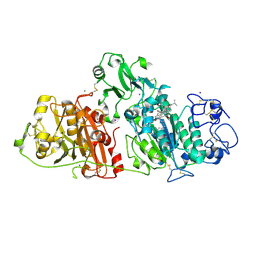 | |
6IAG
 
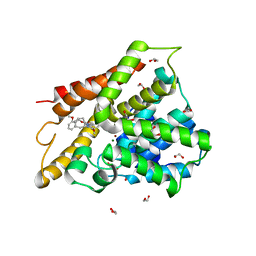 | |
5OTE
 
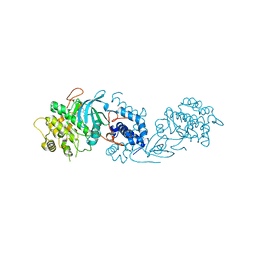 | | MRCK beta in complex with BDP-00008900 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[(6~{S})-1,8-diazaspiro[5.5]undecan-8-yl]-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]-1,3-thiazole, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2017-08-21 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Discovery of Potent and Selective MRCK Inhibitors with Therapeutic Effect on Skin Cancer.
Cancer Res., 78, 2018
|
|
7G9T
 
 | | PanDDA analysis group deposition -- Crystal Structure of Zika virus NS3 Helicase in complex with Z18618496 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Serine protease NS3, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-07-03 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
