1S74
 
 | | SOLUTION STRUCTURE OF A DNA DUPLEX CONTAINING AN ALPHA-ANOMERIC ADENOSINE: INSIGHTS INTO SUBSTRATE RECOGNITION BY ENDONUCLEASE IV | | Descriptor: | 5'-D(*CP*GP*TP*CP*GP*TP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*(A3A)P*CP*GP*AP*CP*G)-3' | | Authors: | Aramini, J.M, Cleaver, S.H, Pon, R.T, Cunningham, R.P, Germann, M.W. | | Deposit date: | 2004-01-28 | | Release date: | 2004-04-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of a DNA Duplex Containing an alpha-Anomeric Adenosine: Insights into Substrate Recognition by Endonuclease IV.
J.Mol.Biol., 338, 2004
|
|
5CLE
 
 | |
5CLV
 
 | | Crystal Structure of KorA-operator DNA complex (KorA-OA) | | Descriptor: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | Authors: | White, S.A, Hyde, E.I, Rajasekar, K.V. | | Deposit date: | 2015-07-16 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
1NZJ
 
 | | Crystal Structure and Activity Studies of Escherichia Coli Yadb ORF | | Descriptor: | Hypothetical protein yadB, ZINC ION | | Authors: | Campanacci, V, Kern, D.Y, Becker, H.D, Spinelli, S, Valencia, C, Vincentelli, R, Pagot, F, Bignon, C, Giege, R, Cambillau, C. | | Deposit date: | 2003-02-18 | | Release date: | 2004-04-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Escherichia coli YadB gene product reveals a novel aminoacyl-tRNA synthetase like activity.
J.Mol.Biol., 337, 2004
|
|
3SF8
 
 | |
3NKD
 
 | | Structure of CRISP-associated protein Cas1 from Escherichia coli str. K-12 | | Descriptor: | CRISPR-associated protein Cas1 | | Authors: | Nocek, B, Skarina, T, Beloglazova, N, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2010-06-18 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair.
Mol.Microbiol., 79, 2011
|
|
1SIQ
 
 | | The Crystal Structure and Mechanism of Human Glutaryl-CoA Dehydrogenase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Glutaryl-CoA dehydrogenase | | Authors: | Wang, M, Fu, Z, Paschke, R, Goodman, S, Frerman, F.E, Kim, J.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Human Glutaryl-CoA Dehydrogenase with and without an Alternate Substrate: Structural Bases of Dehydrogenation and Decarboxylation Reactions
Biochemistry, 43, 2004
|
|
4H7N
 
 | |
7AEW
 
 | |
7AJG
 
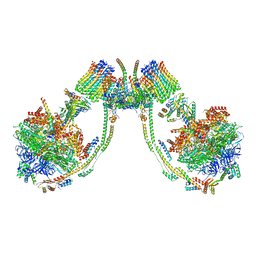 | | bovine ATP synthase dimer state2:state3 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (10.7 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3MZS
 
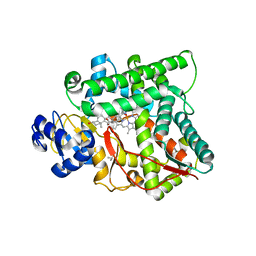 | | Crystal Structure of Cytochrome P450 CYP11A1 in complex with 22-hydroxy-cholesterol | | Descriptor: | (3alpha,8alpha,22R)-cholest-5-ene-3,22-diol, Cholesterol side-chain cleavage enzyme, ISOPROPYL ALCOHOL, ... | | Authors: | Stout, C.D, Annalora, A, Mast, N, Pikuleva, I. | | Deposit date: | 2010-05-12 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Three-step Sequential Catalysis by the Cholesterol Side Chain Cleavage Enzyme CYP11A1.
J.Biol.Chem., 286, 2011
|
|
2WJM
 
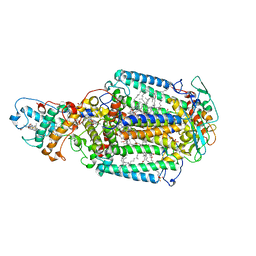 | | Lipidic sponge phase crystal structure of the photosynthetic reaction centre from Blastochloris viridis (low dose) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Woehri, A.B, Wahlgren, W.Y, Malmerberg, E, Johansson, L.C, Neutze, R, Katona, G. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lipidic sponge phase crystal structure of a photosynthetic reaction center reveals lipids on the protein surface.
Biochemistry, 48, 2009
|
|
1AT5
 
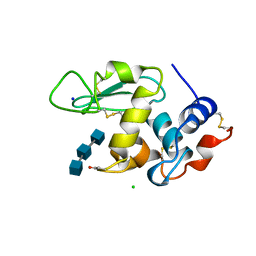 | | HEN EGG WHITE LYSOZYME WITH A SUCCINIMIDE RESIDUE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, LYSOZYME, ... | | Authors: | Noguchi, S, Miyawaki, K, Satow, Y. | | Deposit date: | 1997-08-18 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Succinimide and isoaspartate residues in the crystal structures of hen egg-white lysozyme complexed with tri-N-acetylchitotriose.
J.Mol.Biol., 278, 1998
|
|
4HCX
 
 | |
1B2Y
 
 | |
3N1A
 
 | | Crystal stricture of E145G/Y227F chitinase in complex with cyclo-(L-His-L-Pro) from Bacillus cereus NCTU2 | | Descriptor: | CYCLO-(L-HISTIDINE-L-PROLINE) INHIBITOR, Chitinase A | | Authors: | Hsieh, Y.-C, Wu, Y.-J, Wu, W.-G, Li, Y.-K, Chen, C.-J. | | Deposit date: | 2010-05-15 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of bacillus cereus NCTU2 chitinase complexes with chitooligomers reveal novel substrate binding for catalysis: a chitinase without chitin-binding and insertion domains
J.Biol.Chem., 285, 2010
|
|
4HGF
 
 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGP
 
 | |
3SMV
 
 | | X-ray Crystal Structure of L-Azetidine-2-Carboxylate Hydrolase | | Descriptor: | (S)-2-haloacid dehalogenase, GLYCEROL, IMIDAZOLE | | Authors: | Toyoda, M, Mikami, B, Jitsumori, K, Wackett, L.P, Esaki, N, Kurihara, T. | | Deposit date: | 2011-06-28 | | Release date: | 2012-07-18 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of L-Azetidine-2-carboxylate hydrolase from Pseudomonas sp. strain A2C
To be Published
|
|
3N8Z
 
 | | Crystal Structure of Cyclooxygenase-1 in Complex with Flurbiprofen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FLURBIPROFEN, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
3N0U
 
 | |
4GOA
 
 | |
3N1X
 
 | | X-ray Crystal Structure of Toluene/o-Xylene Monooxygenase Hydroxylase T201C Mutant | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, HYDROXIDE ION, ... | | Authors: | Sazinsky, M.H, McCormick, M.S, Lippard, S.J. | | Deposit date: | 2010-05-17 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Active Site Threonine Facilitates Proton Transfer during Dioxygen Activation at the Diiron Center of Toluene/o-Xylene Monooxygenase Hydroxylase.
J.Am.Chem.Soc., 132, 2010
|
|
1B5Y
 
 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
4HFM
 
 | |
