3SIM
 
 | | Crystallographic structure analysis of family 18 Chitinase from Crocus vernus | | Descriptor: | ACETATE ION, GLYCEROL, Protein, ... | | Authors: | Akrem, A, Iqbal, S, Buck, F, Negm, A, Perbandt, M, Betzel, C. | | Deposit date: | 2011-06-19 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic structure analysis of family 18 Chitinase from Crocus vernus
TO BE PUBLISHED
|
|
1B26
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
3SKK
 
 | | Crystal structure of human arginase I in complex with the inhibitor FABH, Resolution 1.70 A, twinned structure | | Descriptor: | Arginase-1, MANGANESE (II) ION, [(5S)-5-amino-5-carboxy-6,6-difluorohexyl](trihydroxy)borate(1-) | | Authors: | Thorn, K.J, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2011-06-22 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Binding of alpha , alpha-disubstituted amino acids to arginase suggests new avenues for inhibitor design.
J.Med.Chem., 54, 2011
|
|
3NA1
 
 | | Crystal structure of human CYP11A1 in complex with 20-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Adrenodoxin, mitochondrial, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4H4Q
 
 | | Crystal Structure of Ferredoxin reductase, BphA4 E175Q/Q177K (reduced form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
1B6I
 
 | | T4 LYSOZYME MUTANT WITH CYS 54 REPLACED BY THR, CYS 97 REPLACED BY ALA, THR 21 REPLACED BY CYS AND LYS 124 REPLACED BY CYS (C54T,C97A,T21C,K124C) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, PROTEIN (LYSOZYME) | | Authors: | Vetter, I.R, Baase, W.A, Snow, S, Matthews, B.W. | | Deposit date: | 1999-01-14 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Solid-state synthesis and mechanical unfolding of polymers of T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4GWA
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1SMD
 
 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1B7P
 
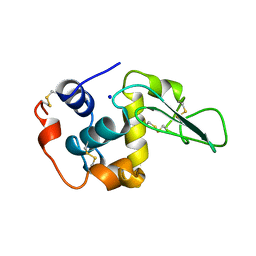 | | VERIFICATION OF SPMP USING MUTANT HUMAN LYSOZYMES | | Descriptor: | PROTEIN (LYSOZYME), SODIUM ION | | Authors: | Takano, K, Ota, M, Ogasahara, K, Yamagata, Y, Nishikawa, K, Yutani, K. | | Deposit date: | 1998-05-08 | | Release date: | 1999-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental verification of the 'stability profile of mutant protein' (SPMP) data using mutant human lysozymes.
Protein Eng., 12, 1999
|
|
3NEP
 
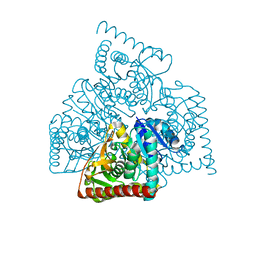 | |
3N8Y
 
 | | Structure of Aspirin Acetylated Cyclooxygenase-1 in Complex with Diclofenac | | Descriptor: | 2-HYDROXYBENZOIC ACID, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sidhu, R.S. | | Deposit date: | 2010-05-28 | | Release date: | 2010-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Comparison of Cyclooxygenase-1 Crystal Structures: Cross-Talk between Monomers Comprising Cyclooxygenase-1 Homodimers
Biochemistry, 49, 2010
|
|
1S9A
 
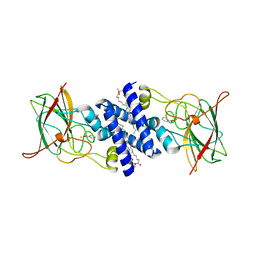 | | Crystal Structure of 4-Chlorocatechol 1,2-dioxygenase from Rhodococcus opacus 1CP | | Descriptor: | (1-HEXADECANOYL-2-TETRADECANOYL-GLYCEROL-3-YL) PHOSPHONYL CHOLINE, BENZOIC ACID, Chlorocatechol 1,2-dioxygenase, ... | | Authors: | Ferraroni, M, Solyanikova, I.P, Kolomytseva, M.P, Scozzafava, A, Golovleva, L.A, Briganti, F. | | Deposit date: | 2004-02-04 | | Release date: | 2004-06-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structure of 4-chlorocatechol 1,2-dioxygenase from the chlorophenol-utilizing gram-positive Rhodococcus opacus 1CP.
J.Biol.Chem., 279, 2004
|
|
4GYL
 
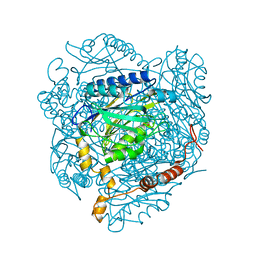 | | The E142L mutant of the amidase from Geobacillus pallidus showing the result of Michael addition of acrylamide at the active site cysteine | | Descriptor: | Aliphatic amidase, CHLORIDE ION, PROPIONAMIDE | | Authors: | Weber, B.W, Sewell, B.T, Kimani, S.W, Varsani, A, Cowan, D.A, Hunter, R. | | Deposit date: | 2012-09-05 | | Release date: | 2013-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The mechanism of the amidases: mutating the glutamate adjacent to the catalytic triad inactivates the enzyme due to substrate mispositioning.
J.Biol.Chem., 288, 2013
|
|
7AJE
 
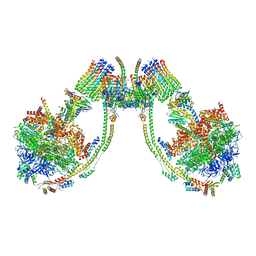 | | bovine ATP synthase dimer state2:state1 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1B8P
 
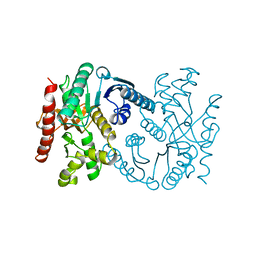 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
4GZQ
 
 | |
3NHK
 
 | |
1B0P
 
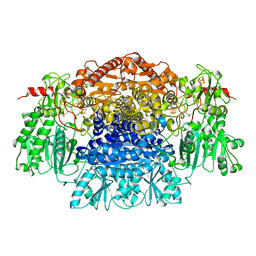 | | CRYSTAL STRUCTURE OF PYRUVATE-FERREDOXIN OXIDOREDUCTASE FROM DESULFOVIBRIO AFRICANUS | | Descriptor: | CALCIUM ION, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Chabriere, E, Charon, M.H, Volbeda, A. | | Deposit date: | 1998-11-12 | | Release date: | 1999-04-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structures of the key anaerobic enzyme pyruvate:ferredoxin oxidoreductase, free and in complex with pyruvate.
Nat.Struct.Biol., 6, 1999
|
|
4H23
 
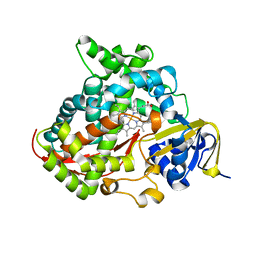 | | Cytochrome P411BM3-CIS cyclopropanation catalyst | | Descriptor: | Cytochrome P450-BM3 variant P411BM3-Cis, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Coelho, P.S, Wang, Z.J, Ener, M.E, Baril, S.A, Kannan, A, Arnold, F.H, Brustad, E.M. | | Deposit date: | 2012-09-11 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A serine-substituted P450 catalyzes highly efficient carbene transfer to olefins in vivo.
Nat.Chem.Biol., 9, 2013
|
|
8W7P
 
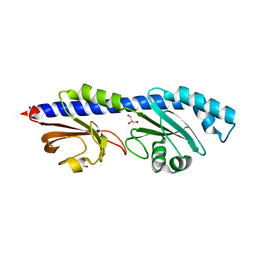 | | Extracellular domain of a sensor histidine kinase | | Descriptor: | Extracellular domain of a sensor histidine kinase NagS, GLYCEROL | | Authors: | Itoh, T, Ogawa, T, Hibi, T, Kimoto, H. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of the extracellular domain of sensor histidine kinase NagS from Paenibacillus sp. str. FPU-7: nagS interacts with oligosaccharide binding protein NagB1 in complexes with N, N'-diacetylchitobiose.
Biosci.Biotechnol.Biochem., 88, 2024
|
|
1B3B
 
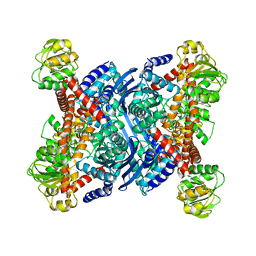 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-07 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
4H4Z
 
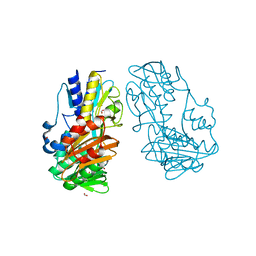 | | Crystal Structure of Ferredoxin reductase, BphA4 E175Q/T176R/Q177G mutant (oxidized form) | | Descriptor: | Biphenyl dioxygenase ferredoxin reductase subunit, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Nishizawa, A, Harada, A, Senda, M, Tachihara, Y, Muramatsu, D, Kishigami, S, Mori, S, Sugiyama, K, Senda, T, Kimura, S. | | Deposit date: | 2012-09-18 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Random Mutagenesis with the Project Assessment for Complete Conversion of Co-Factor Specificity of a Ferredoxin Reductase BphA4
To be Published
|
|
7AJI
 
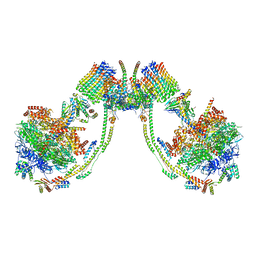 | | bovine ATP synthase dimer state3:state2 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-09-29 | | Release date: | 2021-02-03 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (11.4 Å) | | Cite: | Interface mobility between monomers in dimeric bovine ATP synthase participates in the ultrastructure of inner mitochondrial membranes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
1B74
 
 | | GLUTAMATE RACEMASE FROM AQUIFEX PYROPHILUS | | Descriptor: | D-GLUTAMINE, GLUTAMATE RACEMASE | | Authors: | Hwang, K.Y, Cho, C.S, Kim, S.S, Yu, Y.G, Cho, Y. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of glutamate racemase from Aquifex pyrophilus.
Nat.Struct.Biol., 6, 1999
|
|
1BF6
 
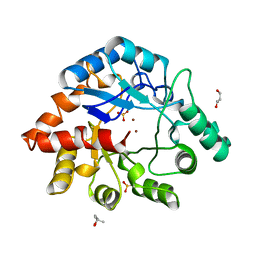 | | PHOSPHOTRIESTERASE HOMOLOGY PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, PHOSPHOTRIESTERASE HOMOLOGY PROTEIN, ... | | Authors: | Buchbinder, J.L, Stephenson, R.C, Scanlan, T.S, Fletterick, R.J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biochemical characterization and crystallographic structure of an Escherichia coli protein from the phosphotriesterase gene family.
Biochemistry, 37, 1998
|
|
