1NOD
 
 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DIMER (DELTA 65) WITH TETRAHYDROBIOPTERIN AND SUBSTRATE L-ARGININE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1998-03-05 | | Release date: | 1999-03-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of nitric oxide synthase oxygenase dimer with pterin and substrate.
Science, 279, 1998
|
|
5BP4
 
 | | Modifying region (DH-ER-KR) of a mycocerosic acid synthase-like (MAS-like) PKS | | Descriptor: | Mycocerosic acid synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Herbst, D.A, Jakob, P.R, Zaehringer, F, Maier, T. | | Deposit date: | 2015-05-27 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Mycocerosic acid synthase exemplifies the architecture of reducing polyketide synthases.
Nature, 531, 2016
|
|
3A2W
 
 | | Peroxiredoxin (C50S) from Aeropytum pernix K1 (peroxide-bound form) | | Descriptor: | GLYCEROL, PEROXIDE ION, Probable peroxiredoxin | | Authors: | Nakamura, T, Kado, Y, Yamaguchi, F, Matsumura, H, Ishikawa, K, Inoue, T. | | Deposit date: | 2009-06-04 | | Release date: | 2009-10-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of peroxiredoxin from Aeropyrum pernix K1 complexed with its substrate, hydrogen peroxide
J.Biochem., 147, 2010
|
|
3EQ0
 
 | | Thrombin Inhibitor | | Descriptor: | (2S)-N-[[2-(aminomethyl)-5-chloro-phenyl]methyl]-1-[(2R)-5-carbamimidamido-2-(phenylmethylsulfonylamino)pentanoyl]pyrrolidine-2-carboxamide, Hirudin variant-1, SODIUM ION, ... | | Authors: | Baum, B, Heine, A, Klebe, G, Steinmetzer, T. | | Deposit date: | 2008-09-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Thrombin Inhibition
To be Published
|
|
8TUM
 
 | |
2RGP
 
 | | Structure of EGFR in complex with hydrazone, a potent dual inhibitor | | Descriptor: | Epidermal growth factor receptor, N-[1-(3-fluorobenzyl)-1H-indazol-5-yl]-5-[(piperidin-1-ylamino)methyl]pyrimidine-4,6-diamine, PHOSPHATE ION | | Authors: | Abad, M.C. | | Deposit date: | 2007-10-04 | | Release date: | 2008-08-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 4-Amino-6-arylamino-pyrimidine-5-carbaldehyde hydrazones as potent ErbB-2/EGFR dual kinase inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4GW1
 
 | | cQFD Meditope | | Descriptor: | Fab heavy chain, Fab light chain, PHOSPHATE ION, ... | | Authors: | Donaldson, J.M, Zer, C, Avery, K.N, Bzymek, K.P, Horne, D.A, Williams, J.C. | | Deposit date: | 2012-08-31 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Identification and grafting of a unique peptide-binding site in the Fab framework of monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5AP3
 
 | | Naturally Occurring Mutations in the MPS1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance. | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, 9-CYCLOPENTYL-2-[[2-METHOXY-4-[(1-METHYLPIPERIDIN-4-YL)OXY]-PHENYL]AMINO]-7-METHYL-7,9-DIHYDRO-8H-PURIN-8-ONE, ... | | Authors: | Gurden, M.D, Westwood, I.M, Faisal, A, Naud, S, Cheung, K.J, McAndrew, C, Wood, A, Schmitt, J, Boxall, K, Mak, G, Workman, P, Burke, R, Hoelder, S, Blagg, J, van Montfort, R.L.M, Linardopoulos, S. | | Deposit date: | 2015-09-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Naturally Occurring Mutations in the Mps1 Gene Predispose Cells to Kinase Inhibitor Drug Resistance.
Cancer Res., 75, 2015
|
|
7PLI
 
 | | DEAD-box helicase DbpA bound to single stranded RNA and ADP/BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DbpA, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Wurm, J.P. | | Deposit date: | 2021-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for RNA-duplex unwinding by the DEAD-box helicase DbpA.
Rna, 29, 2023
|
|
3A4X
 
 | | Crystal structures of catalytic site mutants of active domain 2 of thermostable chitinase from Pyrococcus furiosus complexed with chito-oligosaccharides | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Tsuji, H, Nishimura, S, Inui, T, Ishikawa, K, Nakamura, T, Uegaki, K. | | Deposit date: | 2009-07-22 | | Release date: | 2010-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Kinetic and crystallographic analyses of the catalytic domain of chitinase from Pyrococcus furiosus- the role of conserved residues in the active site
Febs J., 277, 2010
|
|
2DGM
 
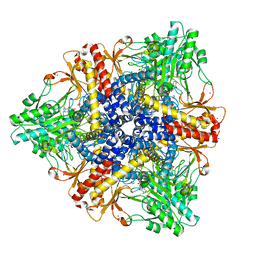 | | Crystal structure of Escherichia coli GadB in complex with iodide | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Gruetter, M.G, Capitani, G, Gut, H. | | Deposit date: | 2006-03-14 | | Release date: | 2006-06-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Escherichia coli acid resistance: pH-sensing, activation by chloride and autoinhibition in GadB
Embo J., 25, 2006
|
|
4GX0
 
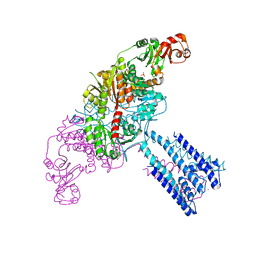 | | Crystal structure of the GsuK L97D mutant | | Descriptor: | CALCIUM ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Kong, C, Zeng, W, Ye, S, Chen, L, Sauer, D.B, Lam, Y, Derebe, M.G, Jiang, Y. | | Deposit date: | 2012-09-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel.
elife, 1, 2012
|
|
7PGA
 
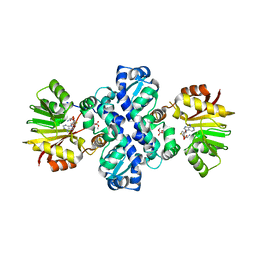 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, S-ADENOSYL-L-HOMOCYSTEINE, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
3EI3
 
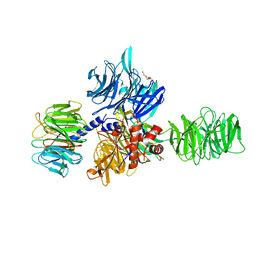 | | Structure of the hsDDB1-drDDB2 complex | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, TETRAETHYLENE GLYCOL | | Authors: | Scrima, A, Thoma, N.H. | | Deposit date: | 2008-09-15 | | Release date: | 2009-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex.
Cell(Cambridge,Mass.), 135, 2008
|
|
3A68
 
 | | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-08-26 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
2ZYF
 
 | | Crystal structure of homocitrate synthase from Thermus thermophilus complexed with magnesuim ion and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, Homocitrate synthase, MAGNESIUM ION | | Authors: | Okada, T, Tomita, T, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanism of substrate recognition and insight into feedback inhibition of homocitrate synthase from thermus thermophilus
J.Biol.Chem., 2009
|
|
1O76
 
 | | CYANIDE COMPLEX OF P450CAM FROM PSEUDOMONAS PUTIDA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CAMPHOR, CYANIDE ION, ... | | Authors: | Fedorov, R, Ghosh, D, Schlichting, I. | | Deposit date: | 2002-10-23 | | Release date: | 2002-12-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Cyanide Complexes of P450Cam and the Oxygenase Domain of Inducible Nitric Oxide Synthase-Structural Models of the Short-Lived Oxygen Complexes
Arch.Biochem.Biophys., 409, 2003
|
|
8TVA
 
 | | Outer Mat-T4P complex | | Descriptor: | Fimbrial protein, Maturation protein | | Authors: | Meng, R, Xing, Z, Thongchol, J, Zhang, J. | | Deposit date: | 2023-08-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (8.55 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
7PXN
 
 | | X-ray structure of LPMO at 6.65x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, COPPER (II) ION | | Authors: | Tandrup, T, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
5EXF
 
 | | Thermostable aldehyde dehydrogenase from Pyrobaculum sp.1860 complexed with NADP+ | | Descriptor: | Aldehyde dehydrogenase, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petrova, T, Bezsudnova, E.Y, Boyko, K.M, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2015-11-23 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | NADP-Dependent Aldehyde Dehydrogenase from ArchaeonPyrobaculum sp.1860: Structural and Functional Features.
Archaea, 2016, 2016
|
|
7KAA
 
 | | NMR solution structures of tirasemtiv drug bound to a fast skeletal troponin C-troponin I complex | | Descriptor: | 6-ethynyl-1-(pentan-3-yl)-1H-imidazo[4,5-b]pyrazin-2-ol, CALCIUM ION, Troponin C, ... | | Authors: | Mercier, P, Li, M.X, Hartman, J.J, Sykes, B.D. | | Deposit date: | 2020-09-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Tirasemtiv Activation of Fast Skeletal Muscle.
J.Med.Chem., 64, 2021
|
|
2ZYS
 
 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
4LYU
 
 | | Fifteen minutes iron loaded frog M ferritin | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Mangani, S, Di Pisa, F, Pozzi, C, Turano, P, Lalli, D. | | Deposit date: | 2013-07-31 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Time-lapse anomalous X-ray diffraction shows how Fe(2+) substrate ions move through ferritin protein nanocages to oxidoreductase sites.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7JUP
 
 | | Structure of human TRPA1 in complex with antagonist compound 21 | | Descriptor: | 1-({3-[(3R,5R)-5-(4-fluorophenyl)oxolan-3-yl]-1,2,4-oxadiazol-5-yl}methyl)-7-methyl-1,7-dihydro-6H-purin-6-one, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L. | | Deposit date: | 2020-08-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Tetrahydrofuran-Based Transient Receptor Potential Ankyrin 1 (TRPA1) Antagonists: Ligand-Based Discovery, Activity in a Rodent Asthma Model, and Mechanism-of-Action via Cryogenic Electron Microscopy.
J.Med.Chem., 64, 2021
|
|
7PE0
 
 | | Crystal structure of IpgC in complex with J52 | | Descriptor: | 2-(1H-imidazol-1-yl)-N-(trans-4-methylcyclohexyl)acetamide, CHLORIDE ION, Chaperone protein IpgC, ... | | Authors: | Gardonyi, M, Heine, A, Klebe, G. | | Deposit date: | 2021-08-09 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of IpgC in complex with J52
To be published
|
|
