3PWM
 
 | | HIV-1 Protease Mutant L76V with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Zhang, Y, Weber, I.T. | | Deposit date: | 2010-12-08 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | The L76V Drug Resistance Mutation Decreases the Dimer Stability and Rate of Autoprocessing of HIV-1 Protease by Reducing Internal Hydrophobic Contacts.
Biochemistry, 50, 2011
|
|
2FS1
 
 | | solution structure of PSD-1 | | Descriptor: | PSD-1 | | Authors: | He, Y, Rozak, D.A, Sari, N, Chen, Y, Bryan, P, Orban, J. | | Deposit date: | 2006-01-20 | | Release date: | 2006-12-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics, and stability variation in bacterial albumin binding modules: implications for species specificity.
Biochemistry, 45, 2006
|
|
1VJK
 
 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
1O0L
 
 | | THE STRUCTURE OF BCL-W REVEALS A ROLE FOR THE C-TERMINAL RESIDUES IN MODULATING BIOLOGICAL ACTIVITY | | Descriptor: | Apoptosis regulator Bcl-W | | Authors: | Hinds, M.G, Lackmann, M, Skea, G.L, Harrison, P.J, Huang, D.C.S, Day, C.L. | | Deposit date: | 2003-02-22 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of Bcl-w reveals a role for the C-terminal residues in modulating biological activity
Embo J., 22, 2003
|
|
3T3G
 
 | | Glycogen Phosphorylase b in complex with GlcBrU | | Descriptor: | 5-bromo-1-(beta-D-glucopyranosyl)pyrimidine-2,4(1H,3H)-dione, Glycogen phosphorylase, muscle form | | Authors: | Kantsadi, A.L, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2011-07-25 | | Release date: | 2012-02-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The sigma-Hole Phenomenon of Halogen Atoms Forms the Structural Basis of the Strong Inhibitory Potency of C5 Halogen Substituted Glucopyranosyl Nucleosides towards Glycogen Phosphorylase b
Chemmedchem, 7, 2012
|
|
2HIG
 
 | | Crystal Structure of Phosphofructokinase apoenzyme from Trypanosoma brucei. | | Descriptor: | 6-phospho-1-fructokinase, SODIUM ION | | Authors: | Martinez-Oyanedel, J, McNae, I.W, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | Deposit date: | 2006-06-29 | | Release date: | 2007-02-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The First Crystal Structure of Phosphofructokinase from a Eukaryote: Trypanosoma brucei.
J.Mol.Biol., 366, 2007
|
|
4DIZ
 
 | |
1W3F
 
 | | Crystal structure of the hemolytic lectin from the mushroom Laetiporus sulphureus complexed with N-acetyllactosamine in the gamma motif | | Descriptor: | GLYCEROL, HEMOLYTIC LECTIN FROM LAETIPORUS SULPHUREUS, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Mancheno, J.M, Tateno, H, Goldstein, I.J, Martinez-Ripoll, M, Hermoso, J.A. | | Deposit date: | 2004-07-15 | | Release date: | 2005-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Analysis of the Laetiporus Sulphureus Hemolytic Pore-Forming Lectin in Complex with Sugars
J.Biol.Chem., 280, 2005
|
|
4P5N
 
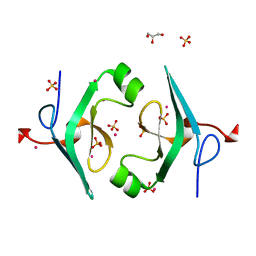 | | Structure of CNAG_02591 from Cryptococcus neoformans | | Descriptor: | CADMIUM ION, GLYCEROL, Hypothetical protein CNAG_02591, ... | | Authors: | Ramagopal, U.A, McClelland, E.E, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-18 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A Small Protein Associated with Fungal Energy Metabolism Affects the Virulence of Cryptococcus neoformans in Mammals.
Plos Pathog., 12, 2016
|
|
4JC1
 
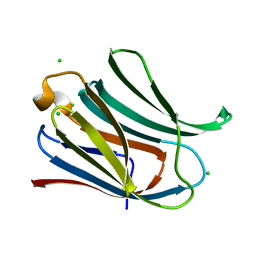 | |
1SBE
 
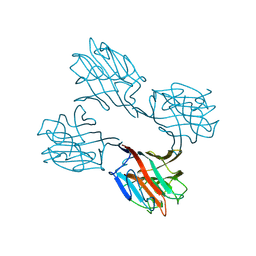 | | SOYBEAN AGGLUTININ FROM GLYCINE MAX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Dessen, A, Olsen, L.R, Gupta, D, Sabesan, S, Sacchettini, J.C, Brewer, C.F. | | Deposit date: | 1995-07-13 | | Release date: | 1998-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin.
Biochemistry, 36, 1997
|
|
4DXB
 
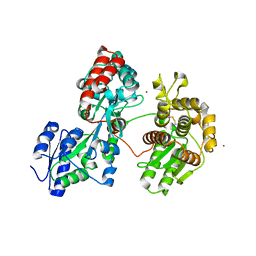 | |
1SCY
 
 | |
1GZP
 
 | | CD1b in complex with GM2 ganglioside | | Descriptor: | B2-MICROGLOBULIN, DOCOSANE, DODECANE, ... | | Authors: | Gadola, S.D, Zaccai, N.R, Harlos, K, Shepherd, D, Ritter, G, Schmidt, R.R, Jones, E.Y, Cerundolo, V. | | Deposit date: | 2002-05-24 | | Release date: | 2002-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains.
Nat. Immunol., 3, 2002
|
|
4JAS
 
 | | Structural basis of a rationally rewired protein-protein interface (HK853mutant A268V, A271G, T275M, V294T and D297E and RR468mutant V13P, L14I, I17M and N21V) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Histidine kinase, MAGNESIUM ION, ... | | Authors: | Podgornaia, A.I, Casino, P, Marina, A, Laub, M.T. | | Deposit date: | 2013-02-19 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of a rationally rewired protein-protein interface critical to bacterial signaling
Structure, 21, 2013
|
|
4PNF
 
 | | Glutathione S-Transferase from Drosophila melanogaster - isozyme E6 | | Descriptor: | GLUTATHIONE, RE21095p | | Authors: | Scian, M, Le Trong, I, Mannervik, B, Atkins, W.M, Stenkamp, R.E. | | Deposit date: | 2014-05-23 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Comparison of epsilon- and delta-class glutathione S-transferases: the crystal structures of the glutathione S-transferases DmGSTE6 and DmGSTE7 from Drosophila melanogaster.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
3VS7
 
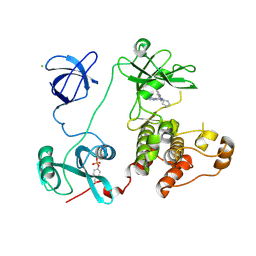 | | Crystal structure of HCK complexed with a pyrazolo-pyrimidine inhibitor 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kuratani, M, Honda, K, Niwa, H, Toyama, M, Handa, N, Yokoyama, S. | | Deposit date: | 2012-04-21 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | A Pyrrolo-Pyrimidine Derivative Targets Human Primary AML Stem Cells in Vivo
Sci Transl Med, 5, 2013
|
|
1NSI
 
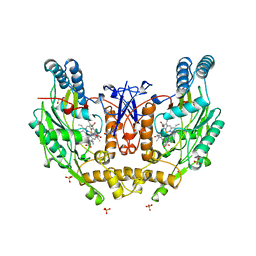 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | Deposit date: | 1999-01-10 | | Release date: | 2000-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
4DDL
 
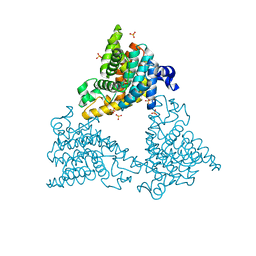 | | PDE10a Crystal Structure Complexed with Novel Inhibitor | | Descriptor: | 2-{1-[5-(6,7-dimethoxycinnolin-4-yl)-3-methylpyridin-2-yl]piperidin-4-yl}propan-2-ol, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S, Zhang, J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of potent, selective, and metabolically stable 4-(pyridin-3-yl)cinnolines as novel phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
1H9D
 
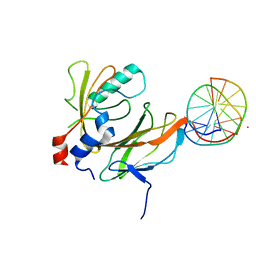 | | Aml1/cbf-beta/dna complex | | Descriptor: | CORE-BINDING FACTOR ALPHA SUBUNIT1, CORE-BINDING FACTOR CBF-BETA, DNA (5'-(*CP*AP*AP*CP*CP*GP*CP*AP*AP*C)-3'), ... | | Authors: | Bravo, J, Warren, A.J. | | Deposit date: | 2001-03-07 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Leukemia-Associated Aml1 (Runx1)-Cbfbeta Complex Functions as a DNA-Induced Molecular Clamp
Nat.Struct.Biol., 8, 2001
|
|
2MBK
 
 | | The Clip-segment of the von Willebrand domain 1 of the BMP modulator protein Crossveinless 2 is preformed | | Descriptor: | Crossveinless 2 | | Authors: | Mueller, T.D, Fiebig, J.E, Weidauer, S.E, Qiu, L, Bauer, M, Schmieder, P, Beerbaum, M, Zhang, J, Oschkinat, H, Sebald, W. | | Deposit date: | 2013-08-02 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The Clip-Segment of the von Willebrand Domain 1 of the BMP Modulator Protein Crossveinless 2 Is Preformed.
Molecules, 18, 2013
|
|
4JOO
 
 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | Descriptor: | (4R)-2'-amino-6-bromo-1',2,2-trimethyl-2,3-dihydrospiro[chromene-4,4'-imidazol]-5'(1'H)-one, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A, Smith, D. | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
2LV1
 
 | | Solution-state NMR structure of prion protein mutant V210I at neutral pH | | Descriptor: | Major prion protein | | Authors: | Biljan, I, Ilc, G, Giachin, G, Legname, G, Plavec, J. | | Deposit date: | 2012-06-26 | | Release date: | 2012-09-19 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Structural Rearrangements at Physiological pH: Nuclear Magnetic Resonance Insights from the V210I Human Prion Protein Mutant.
Biochemistry, 51, 2012
|
|
1S55
 
 | | Mouse RANKL Structure at 1.9A Resolution | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11 | | Authors: | Teale, M.J, Feug, X, Chen, L, Bice, T, Meehan, E.J. | | Deposit date: | 2004-01-19 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Murine RANKL Extra Cellular Domain Homotrimer Structure In Space Groups P212121 And H3 At 1.9 And 2.6 Respectively
To be Published
|
|
4P9Z
 
 | |
