7G9G
 
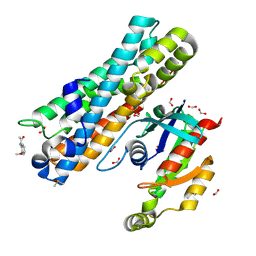 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with PCM-0102236-001 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(2-methoxyphenyl)acetamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
5EL9
 
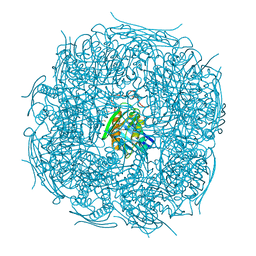 | | A. thaliana IGPD2 in complex with the triazole-phosphonate inhibitor, (S)-C348, to 1.1A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Imidazoleglycerol-phosphate dehydratase 2, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2I1N
 
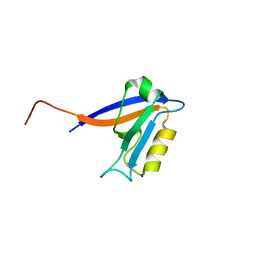 | | Crystal structure of the 1st PDZ domain of Human DLG3 | | Descriptor: | Discs, large homolog 3, SODIUM ION | | Authors: | Turnbull, A.P, Phillips, C, Bunkoczi, G, Debreczeni, J, Ugochukwu, E, Pike, A.C.W, Gorrec, F, Umeano, C, Elkins, J, Berridge, G, Savitsky, P, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
7G9H
 
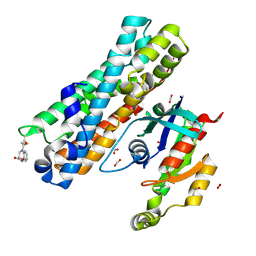 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with PCM-0102245-001 | | Descriptor: | (2S)-4-(chloroacetyl)-3,4-dihydro-2H-1,4-benzoxazine-2-carboxamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G82
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1079512010 | | Descriptor: | 6-methyl-2-[(3-methyl-1,2-oxazol-5-yl)methyl]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
1TPL
 
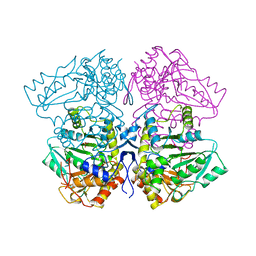 | | THE THREE-DIMENSIONAL STRUCTURE OF TYROSINE PHENOL-LYASE | | Descriptor: | SULFATE ION, TYROSINE PHENOL-LYASE | | Authors: | Antson, A, Demidkina, T, Dauter, Z, Harutyunyan, E, Wilson, K. | | Deposit date: | 1992-11-25 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structure of tyrosine phenol-lyase.
Biochemistry, 32, 1993
|
|
5NIF
 
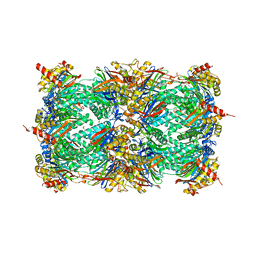 | | Yeast 20S proteasome in complex with Blm-pep activator | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Witkowska, J, Grudnik, P, Golik, P, Dubin, G, Jankowska, E. | | Deposit date: | 2017-03-23 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of a low molecular weight activator Blm-pep with yeast 20S proteasome - insights into the enzyme activation mechanism.
Sci Rep, 7, 2017
|
|
1ASF
 
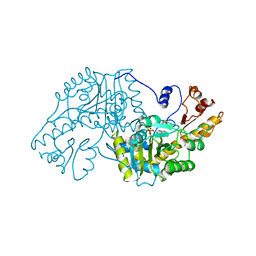 | |
7G83
 
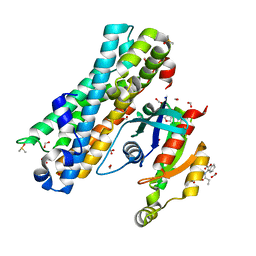 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z108545814 | | Descriptor: | 3-propanamido-1-benzofuran-2-carboxamide, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
7G8Y
 
 | | ARHGEF2 PanDDA analysis group deposition -- ARHGEF2 and RhoA in complex with Z1449748885 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, N-(4-methoxyphenyl)glycinamide, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Bountra, C, von Delft, F, Brennan, P.E. | | Deposit date: | 2023-06-22 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | ARHGEF2 PanDDA analysis group deposition
To Be Published
|
|
4CHK
 
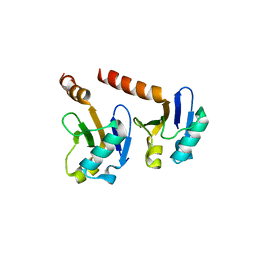 | | Crystal Structure of the ARF5 oligomerization domain | | Descriptor: | AUXIN RESPONSE FACTOR 5 | | Authors: | Nanao, M.H, Mazzoleni, M, Thevenon, E, Brunoud, G, Vernoux, T, Parcy, F, Dumas, R. | | Deposit date: | 2013-12-03 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for Oligomerisation of Auxin Transcriptional Regulators
Nat.Commun., 5, 2014
|
|
5EH4
 
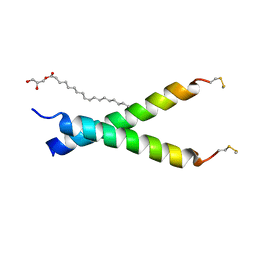 | |
6T9N
 
 | | CryoEM structure of human polycystin-2/PKD2 in UDM supplemented with PI(4,5)P2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wang, Q, Pike, A.C.W, Grieben, M, Baronina, A, Nasrallah, C, Shintre, C, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Lipid Interactions of a Ciliary Membrane TRP Channel: Simulation and Structural Studies of Polycystin-2.
Structure, 28, 2020
|
|
5NKT
 
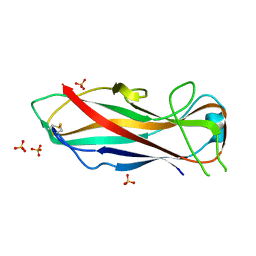 | | FimA wt from E. coli | | Descriptor: | SULFATE ION, Type-1 fimbrial protein, A chain | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2017-04-03 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
7K5N
 
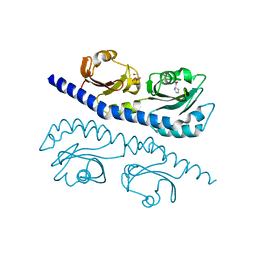 | |
3G3M
 
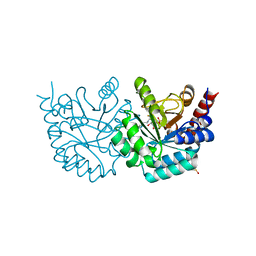 | | Crystal Structure of Human Orotidine 5'-monophosphate Decarboxylase Covalently Modified by 5-fluoro-6-iodo-UMP | | Descriptor: | 5-FLUORO-URIDINE-5'-MONOPHOSPHATE, Uridine 5'-monophosphate synthase | | Authors: | Liu, Y, Tang, H.L, Bello, A.M, Poduch, E, Kotra, L.P, Pai, E.F. | | Deposit date: | 2009-02-02 | | Release date: | 2009-03-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-activity relationships of orotidine-5'-monophosphate decarboxylase inhibitors as anticancer agents.
J.Med.Chem., 52, 2009
|
|
1MJ2
 
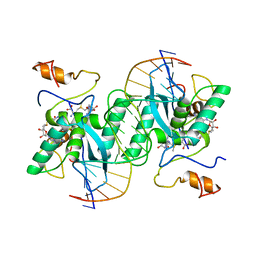 | |
6BWI
 
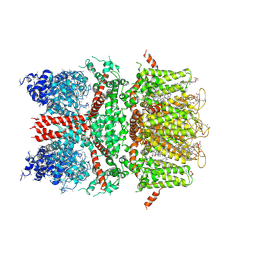 | | 3.7 angstrom cryoEM structure of full length human TRPM4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Zhang, J, Li, Z, Duan, J, Li, J, Clapham, D.E. | | Deposit date: | 2017-12-15 | | Release date: | 2018-12-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of full-length human TRPM4.
Proc.Natl.Acad.Sci.USA, 115, 2018
|
|
5FRM
 
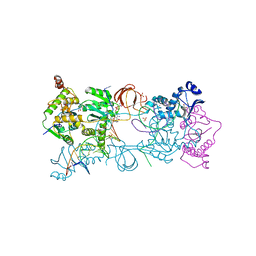 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ384 (compound 4a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-azanylidene-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
4MB5
 
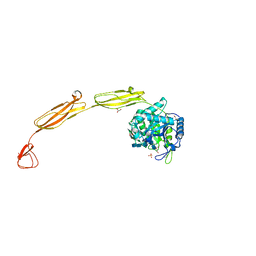 | | Crystal structure of E153Q mutant of cold-adapted chitinase from Moritella complex with Nag5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase 60, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Malecki, P.H, Vorgias, C.E, Rypniewski, W. | | Deposit date: | 2013-08-19 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Crystal structures of substrate-bound chitinase from the psychrophilic bacterium Moritella marina and its structure in solution
Acta Crystallogr.,Sect.D, 70, 2014
|
|
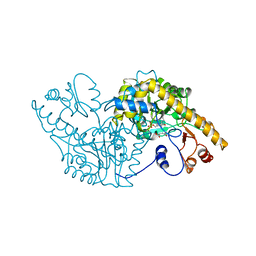
1ASD
 
 | |
3SIL
 
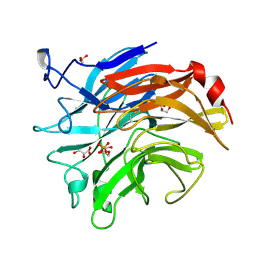 | | SIALIDASE FROM SALMONELLA TYPHIMURIUM | | Descriptor: | GLYCEROL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Garman, E.F, Sheldrick, G.M. | | Deposit date: | 1998-07-07 | | Release date: | 1998-11-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An Enzyme at Atomic Resolution: The 1.05 A Structure of Salmonella Typhimurium Neuraminidase (Sialidase)
To be Published
|
|
7FPM
 
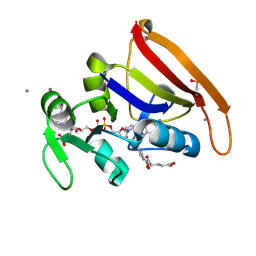 | | DHFR:NADP+:FOL complex at 280 K (crystal 2) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Hekstra, D.R. | | Deposit date: | 2022-09-09 | | Release date: | 2023-09-20 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7FPO
 
 | | DHFR:NADP+:FOL complex at 280 K (crystal 4) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Brookner, D.E, Hekstra, D.R. | | Deposit date: | 2022-09-09 | | Release date: | 2023-09-20 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Perturbative diffraction methods resolve a conformational switch that facilitates a two-step enzymatic mechanism.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1XQA
 
 | |
