2JTV
 
 | | Solution Structure of protein RPA3401, Northeast Structural Genomics Consortium Target RpT7, Ontario Center for Structural Proteomics Target RP3384 | | Descriptor: | Protein of Unknown Function | | Authors: | Ignatchenko, A, Gutmanas, A, Lemak, A, Yee, A, Karra, M, Srisailam, S, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-08-07 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of protein RPA3401, Northeast Structural Genomics Consortium Target RpT7, Ontario Center for Structural Proteomics Target RP3384
TO BE PUBLISHED
|
|
6XX6
 
 | | Arabidopsis thaliana Casein Kinase 2 (CK2) alpha-1 crystal form I | | Descriptor: | CHLORIDE ION, Casein kinase II subunit alpha-1, PIVALIC ACID, ... | | Authors: | Demulder, M, De Veylder, L, Loris, R. | | Deposit date: | 2020-01-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84919691 Å) | | Cite: | Crystal structure of Arabidopsis thaliana casein kinase 2 alpha 1.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2JUY
 
 | | NMR ensemble of Neopetrosiamide A | | Descriptor: | Neopetrosiamide A | | Authors: | Austin, P, Williams, D.E, Heller, M, McIntosh, L.P, Andersen, R.J, Roberge, M, Roskelley, C.D. | | Deposit date: | 2007-09-05 | | Release date: | 2008-08-26 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | NMR ensemble of Neopetrosiamide A
To be Published
|
|
6XX8
 
 | |
2K3W
 
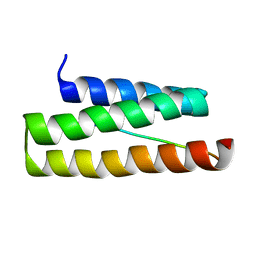 | | NMR structure of VPS4A-MIT-CHMP6 | | Descriptor: | Charged multivesicular body protein 6, Vacuolar protein sorting-associating protein 4A | | Authors: | Kieffer, C, Skalicky, J.J, Morita, E, De Domini, I, Ward, D.M, Kaplan, J, Sundquist, W.I. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding
Dev.Cell, 15, 2008
|
|
2K0P
 
 | |
2JP9
 
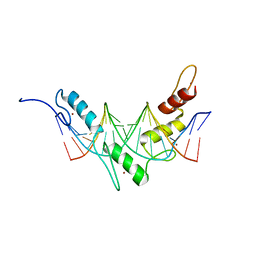 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
8VL3
 
 | |
5NP1
 
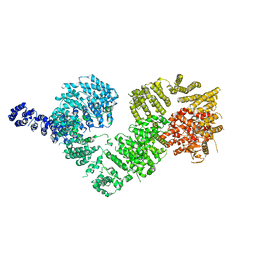 | | Open protomer of human ATM (Ataxia telangiectasia mutated) | | Descriptor: | Serine-protein kinase ATM | | Authors: | Baretic, D, Pollard, H.K, Fisher, D.I, Johnson, C.M, Santhanam, B, Truman, C.M, Kouba, T, Fersht, A.R, Phillips, C, Williams, R.L. | | Deposit date: | 2017-04-13 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of closed and open conformations of dimeric human ATM.
Sci Adv, 3, 2017
|
|
6MKR
 
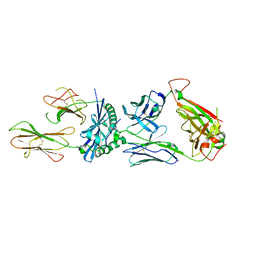 | | 5287 TCR bound to IAb Padi4 | | Descriptor: | 5287 TCR alpha chain, 5287 TCR beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Blevins, S.J, Huseby, E.S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.355 Å) | | Cite: | A temporal thymic selection switch and ligand binding kinetics constrain neonatal Foxp3+Tregcell development.
Nat.Immunol., 20, 2019
|
|
2JI3
 
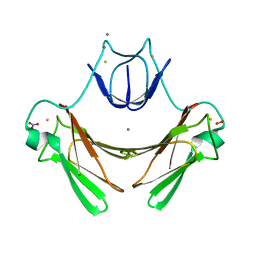 | | X-ray structure of the iron-peroxide intermediate of superoxide reductase (E114A mutant) from Desulfoarculus baarsii | | Descriptor: | CALCIUM ION, Desulfoferrodoxin, FE (III) ION, ... | | Authors: | Katona, G, Carpentier, P, Niviere, V, Amara, P, Adam, V, Ohana, J, Tsanov, N, Bourgeois, D. | | Deposit date: | 2007-02-24 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme.
Science, 316, 2007
|
|
2JEO
 
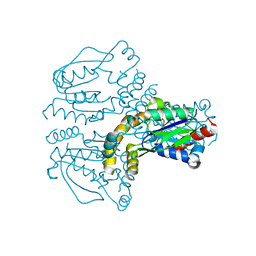 | | Crystal structure of human uridine-cytidine kinase 1 | | Descriptor: | URIDINE-CYTIDINE KINASE 1 | | Authors: | Kosinska, U, Stenmark, P, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg Schiavone, L, Hogbom, M, Johansson, I, Karlberg, T, Kotenyova, T, Moche, M, Nilsson, M.E.P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Uppsten, M, Thorsell, A.G, van den Berg, S, Weigelt, J, Welin, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Human Uridine-Cytidine Kinase 1
To be Published
|
|
2MAF
 
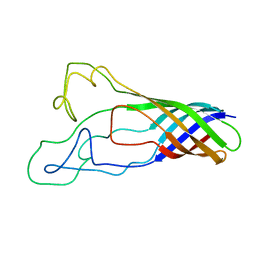 | | Solution structure of opa60 from n. gonorrhoeae | | Descriptor: | Opacity protein opA60 | | Authors: | Columbus, L, Fox, D.A, Larsson, P, Lo, R.H, Kroncke, B.M, Kasson, P.M. | | Deposit date: | 2013-07-08 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neisserial outer membrane protein Opa60: loop flexibility essential to receptor recognition and bacterial engulfment.
J.Am.Chem.Soc., 136, 2014
|
|
8VL4
 
 | |
2MF8
 
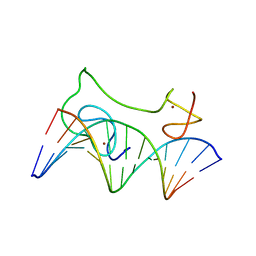 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
2M8N
 
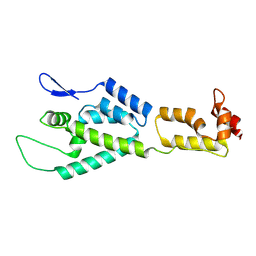 | | HIV-1 capsid monomer structure | | Descriptor: | Capsid protein p24 | | Authors: | Deshmukh, L, Schwieters, C.D, Grishaev, A, Clore, G, Ghirlando, R. | | Deposit date: | 2013-05-24 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Structure and Dynamics of Full-Length HIV-1 Capsid Protein in Solution.
J.Am.Chem.Soc., 135, 2013
|
|
2MAG
 
 | | NMR STRUCTURE OF MAGAININ 2 IN DPC MICELLES, 10 STRUCTURES | | Descriptor: | MAGAININ 2 | | Authors: | Gesell, J.J, Zasloff, M, Opella, S.J. | | Deposit date: | 1997-12-19 | | Release date: | 1998-04-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Two-dimensional 1H NMR experiments show that the 23-residue magainin antibiotic peptide is an alpha-helix in dodecylphosphocholine micelles, sodium dodecylsulfate micelles, and trifluoroethanol/water solution.
J.Biomol.NMR, 9, 1997
|
|
2NOS
 
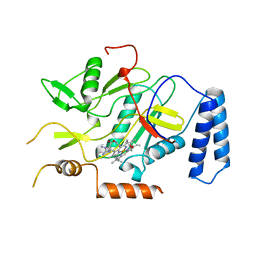 | | MURINE INDUCIBLE NITRIC OXIDE SYNTHASE OXYGENASE DOMAIN (DELTA 114), AMINOGUANIDINE COMPLEX | | Descriptor: | AMINOGUANIDINE, IMIDAZOLE, INDUCIBLE NITRIC OXIDE SYNTHASE, ... | | Authors: | Crane, B.R, Arvai, A.S, Getzoff, E.D, Stuehr, D.J, Tainer, J.A. | | Deposit date: | 1997-09-28 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of nitric oxide synthase oxygenase domain and inhibitor complexes.
Science, 278, 1997
|
|
5NCE
 
 | | Structure of PsDef1 defensin from Pinus sylvestris | | Descriptor: | Defensin-1 | | Authors: | Khairutdinov, B.I, Ermakova, E.A, Bessolitsyna, E.K, Toporkova, Y.Y, Tarasova, N.B, Kovaleva, V, Zuev, Y.F, Nesmelova, I.V. | | Deposit date: | 2017-03-03 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR structure, conformational dynamics, and biological activity of PsDef1 defensin from Pinus sylvestris.
Biochim. Biophys. Acta, 1865, 2017
|
|
2MUV
 
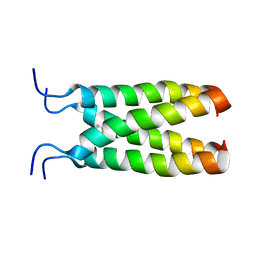 | | NOE-based model of the influenza A virus M2 (19-49) bound to drug 11 | | Descriptor: | (3s,5s,7s)-N-[(5-bromothiophen-2-yl)methyl]tricyclo[3.3.1.1~3,7~]decan-1-aminium, Matrix protein 2 | | Authors: | Wu, Y, Wang, J, DeGrado, W. | | Deposit date: | 2014-09-18 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Flipping in the Pore: Discovery of Dual Inhibitors That Bind in Different Orientations to the Wild-Type versus the Amantadine-Resistant S31N Mutant of the Influenza A Virus M2 Proton Channel.
J.Am.Chem.Soc., 136, 2014
|
|
5NAM
 
 | |
2MT6
 
 | | Solution structure of the human ubiquitin conjugating enzyme Ube2w | | Descriptor: | Ubiquitin-conjugating enzyme E2 W | | Authors: | Vittal, V, Shi, L, Wenzel, D.M, Brzovic, P.S, Klevit, R.E. | | Deposit date: | 2014-08-14 | | Release date: | 2014-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intrinsic disorder drives N-terminal ubiquitination by Ube2w.
Nat.Chem.Biol., 11, 2015
|
|
2N04
 
 | |
2MRE
 
 | | NMR structure of the Rad18-UBZ/ubiquitin complex | | Descriptor: | E3 ubiquitin-protein ligase RAD18, Polyubiquitin-C, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2MXU
 
 | | 42-Residue Beta Amyloid Fibril | | Descriptor: | Amyloid beta A4 protein | | Authors: | Xiao, Y, Ma, B, McElheny, D, Parthasarathy, S, Long, F, Hoshi, M, Nussinov, R, Ishii, Y. | | Deposit date: | 2015-01-14 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A beta (1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
