7P6J
 
 | | Crystal structure of glycosyl-enzyme intermediate of RBcel1 Y201F | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Collet, L, Dutoit, R. | | Deposit date: | 2021-07-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Highlighting the factors governing transglycosylation in the GH5_5 endo-1,4-beta-glucanase RBcel1.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7AKM
 
 | | Crystal structure of CHK1 kinase domain in complex with ATPyS | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Day, M, Oliver, A.W, Pearl, L.H. | | Deposit date: | 2020-10-01 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for recruitment of the CHK1 DNA damage kinase by the CLASPIN scaffold protein.
Structure, 29, 2021
|
|
6DU2
 
 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | MAGNESIUM ION, REST-pS861/4, carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
2VZI
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD4 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin,Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
7AWF
 
 | | The Fk1 domain of FKBP51 in complex with (2R,5S,12R)-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^5,^10]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone | | Descriptor: | (2~{R},5~{S},12~{R})-12-cyclohexyl-2-[2-(3,4-dimethoxyphenyl)ethyl]-15,15,16-trimethyl-3,19-dioxa-10,13,16-triazatricyclo[18.3.1.0^{5,10}]tetracosa-1(24),20,22-triene-4,11,14,17-tetrone, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Voll, M.A, Meyners, C, Heymann, T, Merz, S, Purder, P, Bracher, A, Hausch, F. | | Deposit date: | 2020-11-07 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Macrocyclic FKBP51 Ligands Define a Transient Binding Mode with Enhanced Selectivity.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
2XEN
 
 | | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins With a Redesigned C-capping Module. | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, NI1C MUT4, ... | | Authors: | Kramer, M, Wetzel, S.K, Pluckthun, A, Mittl, P, Grutter, M. | | Deposit date: | 2010-05-17 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Determinants for Improved Thermal Stability of Designed Ankyrin Repeat Proteins with a Redesigned C-Capping Module.
J.Mol.Biol., 404, 2010
|
|
9GOX
 
 | | Crystal structure of Fab B6-D9 in complex with CD38 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Dreyfus, C, Freier, R. | | Deposit date: | 2024-09-06 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biparatopic binding of ISB 1442 to CD38 in trans enables increased cell antibody density and increased avidity.
Mabs, 17, 2025
|
|
6DCL
 
 | | Crystal structure of UP1 bound to pri-miRNA-18a terminal loop | | Descriptor: | 1,2-ETHANEDIOL, Heterogeneous nuclear ribonucleoprotein A1, RNA (5'-R(*AP*GP*UP*AP*GP*AP*UP*UP*AP*GP*C)-3') | | Authors: | Kooshapur, H, Sattler, M. | | Deposit date: | 2018-05-07 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Structural basis for terminal loop recognition and stimulation of pri-miRNA-18a processing by hnRNP A1.
Nat Commun, 9, 2018
|
|
5CYU
 
 | | Structure of the soluble domain of EccB1 from the Mycobacterium smegmatis ESX-1 secretion system. | | Descriptor: | Conserved membrane protein | | Authors: | Arbing, M.A, Chan, S, Kahng, S, Kim, J, Eisenberg, D.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-30 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structures of EccB1 and EccD1 from the core complex of the mycobacterial ESX-1 type VII secretion system.
Bmc Struct.Biol., 16, 2016
|
|
4RME
 
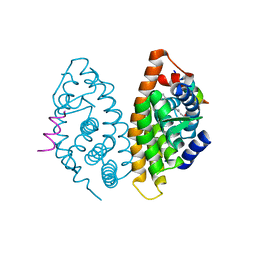 | | Crystal structure of human Retinoid X receptor alpha ligand binding domain complex with 9cUAB111 and coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-[2-(3-methylbutyl)-3-(propan-2-yl)cyclohex-2-en-1-ylidene]octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Muccio, D.D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conformationally Defined Rexinoids and Their Efficacy in the Prevention of Mammary Cancers.
J.Med.Chem., 58, 2015
|
|
8S1X
 
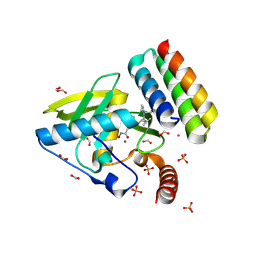 | | Crystal structure of Actinonin-bound PDF1 and the computationally designed DBAct553_1 protein binder | | Descriptor: | ACTINONIN, DBAct553_1, FORMIC ACID, ... | | Authors: | Marchand, A, Pacesa, M, Correia, B.E. | | Deposit date: | 2024-02-16 | | Release date: | 2024-10-30 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Targeting protein-ligand neosurfaces with a generalizable deep learning tool.
Nature, 639, 2025
|
|
6GRO
 
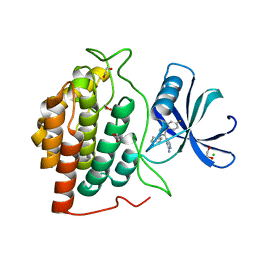 | | Human CSNK1G3 bound to SB-223133 | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(2-azanylpyrimidin-4-yl)-4-(4-fluorophenyl)imidazol-1-yl]cyclohexan-1-ol, CHLORIDE ION, ... | | Authors: | Szklarz, M, Vollmar, M, von Delft, F, Bountra, C, Knapp, S, Edwards, A.M, Arrowsmith, C, Elkins, J.M. | | Deposit date: | 2018-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | CSNK1G3 bound to SB-223133
To Be Published
|
|
6DPG
 
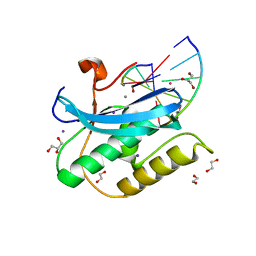 | |
6ZIY
 
 | | Respiratory complex I from Thermus thermophilus, NADH dataset, major state | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Kaszuba, K, Tambalo, M, Gallagher, G.T, Sazanov, L.A. | | Deposit date: | 2020-06-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
5G3P
 
 | | Bacillus cereus formamidase (BceAmiF) acetylated at the active site. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gavira, J.A, Conejero-Muriel, M, Martinez-Rodriguez, S. | | Deposit date: | 2016-04-29 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Arch.Biochem.Biophys., 662, 2019
|
|
6GVG
 
 | | Crystal structure of PI3K alpha in complex with 3-(2-Amino-benzooxazol-5-yl)-1-isopropyl-4-methyl-1H-pyrazolo[3,4-d]pyrimidin-6-ylamine | | Descriptor: | 5-(6-azanyl-4-methyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzoxazol-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Ouvry, G, Aurelly, M, Bonnary, L, Borde, E, Bouix-Peter, C, Chantalat, L, Clary, L, Defoin-Platel, C, Deret, S, Forissier, M, Harris, C.S, Isabet, T, Lamy, L, Luzy, A.P, Pascau, J, Soulet, C, Taddei, A, Taquet, N, Tomas, L, Thoreau, E, Varvier, E, Vial, E, Hennequin, L.F. | | Deposit date: | 2018-06-21 | | Release date: | 2019-10-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Impact of Minor Structural Modifications on Properties of a Series of mTOR Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
7MLC
 
 | | PYL10 bound to the ABA pan-antagonist 4a | | Descriptor: | 1-{2-[3,5-dicyclopropyl-4-(4-{[(quinoxaline-2-carbonyl)amino]methyl}-1H-1,2,3-triazol-1-yl)phenyl]acetamido}cyclohexane-1-carboxylic acid, Abscisic acid receptor PYL10, GLYCEROL | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
9GPE
 
 | |
8S2N
 
 | |
6BBV
 
 | | Crystal Structure of JAK2 in complex with compound 25 | | Descriptor: | N-{cis-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Han, S. | | Deposit date: | 2017-10-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of N-{cis-3-[Methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]cyclobutyl}propane-1-sulfonamide (PF-04965842): A Selective JAK1 Clinical Candidate for the Treatment of Autoimmune Diseases.
J. Med. Chem., 61, 2018
|
|
5KH5
 
 | |
7UK1
 
 | |
8I5T
 
 | |
6DP6
 
 | |
5CZ0
 
 | |
