1VFF
 
 | |
1VFG
 
 | | Crystal structure of tRNA nucleotidyltransferase complexed with a primer tRNA and an incoming ATP analog | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, RNA (75-MER), poly A polymerase | | Authors: | Tomita, K, Fukai, S, Ishitani, R, Ueda, T, Takeuchi, N, Vassylyev, D.G, Nureki, O, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for template-independent RNA polymerization.
Nature, 430, 2004
|
|
1VFH
 
 | | Crystal structure of alanine racemase from D-cycloserine producing Streptomyces lavendulae | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFI
 
 | | Solution Structure of Vanabin2 (RUH-017), a Vanadium-binding Protein from Ascidia sydneiensis samea | | Descriptor: | vanadium-binding protein 2 | | Authors: | Hamada, T, Hirota, H, Asanuma, M, Hayashi, F, Kobayashi, N, Ueki, T, Michibata, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-13 | | Release date: | 2005-03-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vanabin2, a Vanadium(IV)-Binding Protein from the Vanadium-Rich Ascidian Ascidia sydneiensis samea
J.Am.Chem.Soc., 127, 2005
|
|
1VFJ
 
 | | Crystal structure of TT1020 from Thermus thermophilus HB8 | | Descriptor: | nitrogen regulatory protein p-II | | Authors: | Wang, H, Sakai, H, Takemoto-Hori, C, Kaminishi, T, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-04-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the signal transducing protein GlnK from Thermus thermophilus HB8
J.STRUCT.BIOL., 149, 2005
|
|
1VFL
 
 | | Adenosine deaminase | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2004-04-16 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Compound Recognition by Adenosine Deaminase
Biochemistry, 44, 2005
|
|
1VFM
 
 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/alpha-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFN
 
 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | HYPOXANTHINE, MAGNESIUM ION, PURINE-NUCLEOSIDE PHOSPHORYLASE, ... | | Authors: | Koellner, G, Bzowska, A. | | Deposit date: | 1996-10-08 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of calf spleen purine nucleoside phosphorylase in a complex with hypoxanthine at 2.15 A resolution.
J.Mol.Biol., 265, 1997
|
|
1VFO
 
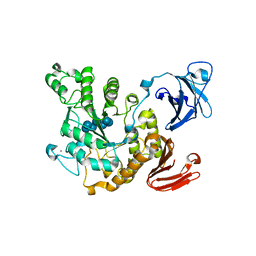 | | Crystal structure of Thermoactinomyces vulgaris R-47 alpha-amylase 2/beta-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclic alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Cycloheptakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-16 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFP
 
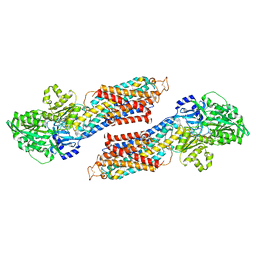 | |
1VFQ
 
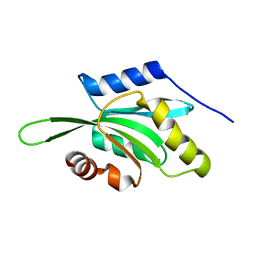 | |
1VFR
 
 | | THE MAJOR NAD(P)H:FMN OXIDOREDUCTASE FROM VIBRIO FISCHERI | | Descriptor: | FLAVIN MONONUCLEOTIDE, NAD(P)H:FMN OXIDOREDUCTASE | | Authors: | Koike, H, Sasaki, H, Kobori, T, Zenno, S, Saigo, K, Murphy, M.E.P, Adman, E.T, Tanokura, M. | | Deposit date: | 1998-01-09 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 A crystal structure of the major NAD(P)H:FMN oxidoreductase of a bioluminescent bacterium, Vibrio fischeri: overall structure, cofactor and substrate-analog binding, and comparison with related flavoproteins.
J.Mol.Biol., 280, 1998
|
|
1VFS
 
 | | Crystal structure of D-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | Descriptor: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFT
 
 | | Crystal structure of L-cycloserine-bound form of alanine racemase from D-cycloserine-producing Streptomyces lavendulae | | Descriptor: | CHLORIDE ION, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, alanine racemase | | Authors: | Noda, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2004-04-19 | | Release date: | 2004-09-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product.
J.Biol.Chem., 279, 2004
|
|
1VFU
 
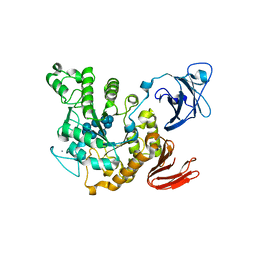 | | Crystal structure of Thermoactinomyces vulgaris R-47 amylase 2/gamma-cyclodextrin complex | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), Neopullulanase 2 | | Authors: | Ohtaki, A, Mizuno, M, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2004-04-19 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Complex structures of Thermoactinomyces vulgaris R-47 alpha-amylase 2 with acarbose and cyclodextrins demonstrate the multiple substrate recognition mechanism
J.BIOL.CHEM., 279, 2004
|
|
1VFV
 
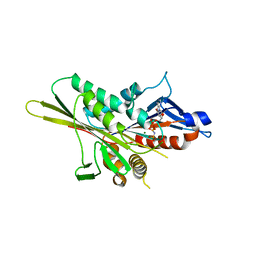 | | Crystal Structure of the Kif1A Motor Domain Complexed With Mg-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (Fusion protein consisting of Kinesin-like protein KIF1A, ... | | Authors: | Nitta, R, Kikkawa, M, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-04-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | KIF1A Alternately Uses Two Loops to Bind Microtubules
Science, 305, 2004
|
|
1VFW
 
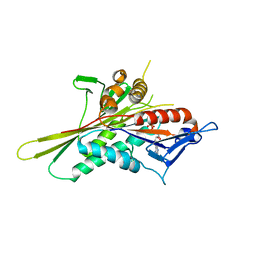 | | Crystal Structure of the Kif1A Motor Domain Complexed With Mg-AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (Fusion protein consisting of Kinesin-like protein KIF1A, ... | | Authors: | Nitta, R, Kikkawa, M, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-04-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | KIF1A Alternately Uses Two Loops to Bind Microtubules
Science, 305, 2004
|
|
1VFX
 
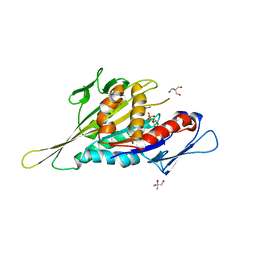 | | Crystal Structure of the Kif1A Motor Domain Complexed With ADP-Mg-AlFx | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Nitta, R, Kikkawa, M, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-04-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | KIF1A Alternately Uses Two Loops to Bind Microtubules
Science, 305, 2004
|
|
1VFY
 
 | |
1VFZ
 
 | | Crystal Structure of the Kif1A Motor Domain Complexed With ADP-Mg-VO4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (Fusion protein consisting of Kinesin-like protein KIF1A, ... | | Authors: | Nitta, R, Kikkawa, M, Okada, Y, Hirokawa, N. | | Deposit date: | 2004-04-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | KIF1A Alternately Uses Two Loops to Bind Microtubules
Science, 305, 2004
|
|
1VG0
 
 | | The crystal structures of the REP-1 protein in complex with monoprenylated Rab7 protein | | Descriptor: | CHLORIDE ION, GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
1VG1
 
 | | GDP-Bound Rab7 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7 | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
1VG2
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG3
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima A76Y/S77F mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
1VG4
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima F132A/L128A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Ko, T.P, Chou, C.C, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A molecular ruler for chain elongation catalyzed by octaprenyl pyrophosphate synthase and its structure-based engineering to produce unprecedented long chain trans-prenyl products
Biochemistry, 43, 2004
|
|
