8J58
 
 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in the apo-form | | Descriptor: | ATP synthase subunit a, ATP synthase subunit c | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8J0S
 
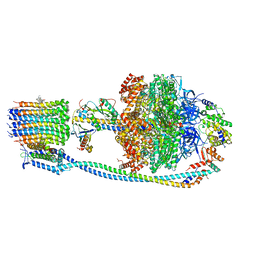 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in complex with bedaquiline(BDQ) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8JR0
 
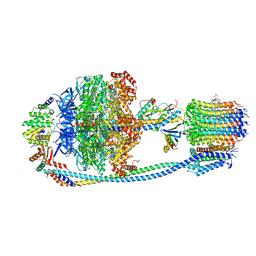 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
8JR1
 
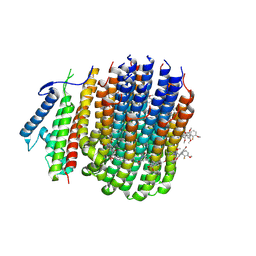 | | Cryo-EM structure of Mycobacterium tuberculosis ATP synthase Fo in complex with TBAJ-587 | | Descriptor: | (1~{S},2~{S})-1-(6-bromanyl-2-methoxy-quinolin-3-yl)-2-(2,6-dimethoxypyridin-4-yl)-4-(dimethylamino)-1-(2-fluoranyl-3-methoxy-phenyl)butan-2-ol, ATP synthase subunit a, ATP synthase subunit c | | Authors: | Zhang, Y, Lai, Y, Liu, F, Rao, Z, Gong, H. | | Deposit date: | 2023-06-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Inhibition of M. tuberculosis and human ATP synthase by BDQ and TBAJ-587.
Nature, 631, 2024
|
|
3LG8
 
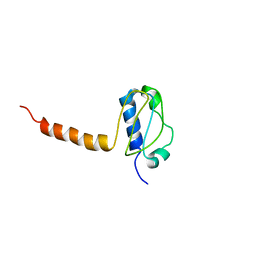 | | Crystal structure of the C-terminal part of subunit E (E101-206) from Methanocaldococcus jannaschii of A1AO ATP synthase | | Descriptor: | A-type ATP synthase subunit E | | Authors: | Balakrishna, A.M, Manimekalai, M.S.S, Hunke, C, Gayen, S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-01-19 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Crystal and solution structure of the C-terminal part of the Methanocaldococcus jannaschii A1AO ATP synthase subunit E revealed by X-ray diffraction and small-angle X-ray scattering
J.Bioenerg.Biomembr., 42, 2010
|
|
3MFY
 
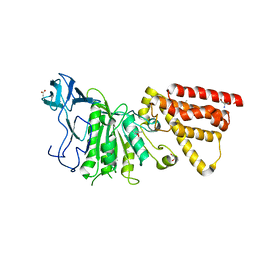 | | Structural characterization of the subunit A mutant F236A of the A-ATP synthase from Pyrococcus horikoshii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Balakrishna, A.M, Kumar, A, Manimekali, M.S.S, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-04-05 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
7JG6
 
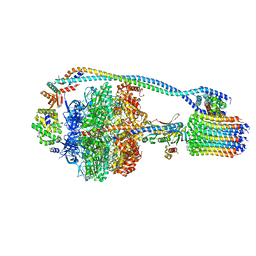 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
3M4Y
 
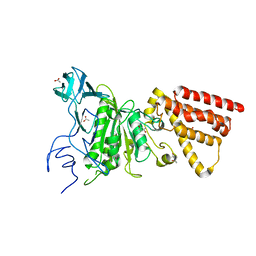 | | Structural characterization of the subunit A mutant P235A of the A-ATP synthase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ... | | Authors: | Manimekalai, M.S, Balakrishna, A.M, Kumar, A, Priya, R, Biukovic, G, Jeyakanthan, J, Gruber, G. | | Deposit date: | 2010-03-12 | | Release date: | 2011-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The critical roles of residues P235 and F236 of subunit A of the motor protein A-ATP synthase in P-loop formation and nucleotide binding.
J.Mol.Biol., 401, 2010
|
|
8CZ7
 
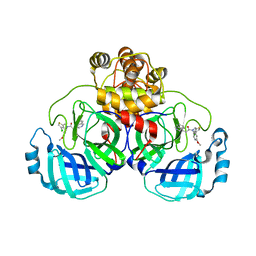 | | Crystal structure of SARS-CoV-2 Mpro with compound C2 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-(4-methoxyphenyl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYZ
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C4 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)-N-[4-(methylsulfanyl)phenyl]acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CYU
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C5 | | Descriptor: | 3C-like proteinase, N-[(4-chlorothiophen-2-yl)methyl]-N-[4-(dimethylamino)phenyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
8CZ4
 
 | | Crystal structure of SARS-CoV-2 Mpro with compound C3 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-N-[(4-chlorothiophen-2-yl)methyl]-2-(isoquinolin-4-yl)acetamide | | Authors: | Worrall, L.J, Lee, J, Strynadka, N.C.J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants.
Emerg Microbes Infect, 12, 2023
|
|
7JG7
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 3 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGB
 
 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG8
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 1 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
8IGU
 
 | |
8IGW
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
7JGA
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase rotational state 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG5
 
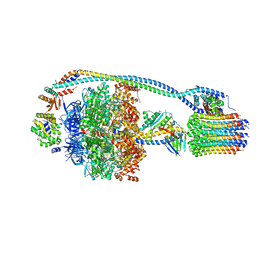 | | Cryo-EM structure of bedaquiline-free Mycobacterium smegmatis ATP synthase rotational state 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JGC
 
 | | Cryo-EM structure of bedaquiline-saturated Mycobacterium smegmatis ATP synthase FO region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit b-delta, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
7JG9
 
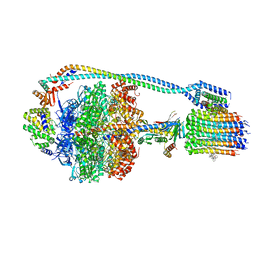 | | Cryo-EM structure of bedaquiline-saturated mycobacterium smegmatis ATP synthase rotational state 2 (backbone model) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Guo, H, Courbon, G.M, Rubinstein, J.L. | | Deposit date: | 2020-07-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of mycobacterial ATP synthase bound to the tuberculosis drug bedaquiline.
Nature, 589, 2021
|
|
2KK7
 
 | |
6J5I
 
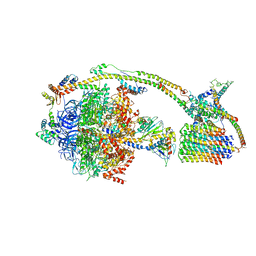 | | Cryo-EM structure of the mammalian DP-state ATP synthase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase F1 subunit epsilon, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
6J54
 
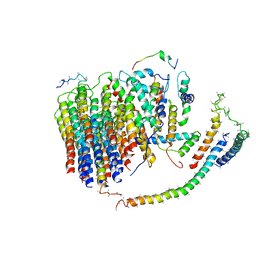 | | Cryo-EM structure of the mammalian E-state ATP synthase FO section | | Descriptor: | ATP synthase membrane subunit 6.8PL, ATP synthase membrane subunit DAPIT, ATP synthase peripheral stalk-membrane subunit b, ... | | Authors: | Gu, J, Zhang, L, Yi, J, Yang, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Cryo-EM structure of the mammalian ATP synthase tetramer bound with inhibitory protein IF1.
Science, 364, 2019
|
|
