4XSX
 
 | | Crystal structure of CBR 703 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.708 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7VF9
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
4XLP
 
 | |
4XSY
 
 | | Crystal structure of CBR 9379 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 3-{[(2,6-dichlorophenyl)carbamoyl]amino}-N-hydroxy-N'-phenyl-5-(trifluoromethyl)benzenecarboximidamide, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.007 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7W5W
 
 | |
7W5X
 
 | | Cryo-EM structure of SoxS-dependent transcription activation complex with zwf promoter DNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Feng, Y, Shi, J. | | Deposit date: | 2021-11-30 | | Release date: | 2022-10-26 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of three different transcription activation strategies adopted by a single regulator SoxS.
Nucleic Acids Res., 50, 2022
|
|
4YLO
 
 | | E. coli Transcription Initiation Complex - 16-bp spacer and 4-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YFN
 
 | | Escherichia coli RNA polymerase in complex with squaramide compound 14 (N-[3,4-dioxo-2-(4-{[4-(trifluoromethyl)benzyl]amino}piperidin-1-yl)cyclobut-1-en-1-yl]-3,5-dimethyl-1,2-oxazole-4-sulfonamide) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Molodtsov, V, Fleming, P.R, Eyermann, C.J, Ferguson, A.D, Foulk, M.A, McKinney, D.C, Masse, C.E, Buurman, E.T, Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.817 Å) | | Cite: | X-ray Crystal Structures of Escherichia coli RNA Polymerase with Switch Region Binding Inhibitors Enable Rational Design of Squaramides with an Improved Fraction Unbound to Human Plasma Protein.
J.Med.Chem., 58, 2015
|
|
4ZH3
 
 | |
4ZH4
 
 | | Crystal structure of Escherichia coli RNA polymerase in complex with CBRP18 | | Descriptor: | 5-(4-fluorophenyl)-4-[4-fluoro-3-(trifluoromethyl)phenyl]-1H-pyrazole, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Feng, Y, Ebright, R.H. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.993 Å) | | Cite: | Structural Basis of Transcription Inhibition by CBR Hydroxamidines and CBR Pyrazoles.
Structure, 23, 2015
|
|
7XL4
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (closed lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7XL3
 
 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes with transcription factor SutA (open lobe) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2022-04-21 | | Release date: | 2022-07-27 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
7X76
 
 | |
7X74
 
 | |
7X75
 
 | |
4ZH2
 
 | |
4XLR
 
 | |
4XLN
 
 | |
7YE1
 
 | | The cryo-EM structure of C. crescentus GcrA-TACup | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (57-MER)-non template, DNA (57-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7YE2
 
 | | The cryo-EM structure of C. crescentus GcrA-TACdown | | Descriptor: | Cell cycle regulatory protein GcrA, DNA (90-MER)-non template, DNA (90-MER)-template, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of Caulobacter crescentus transcription activation complex with an essential cell cycle regulator GcrA
Nucleic Acids Res., 2023
|
|
7W5Y
 
 | |
7VPD
 
 | |
7VPZ
 
 | |
4YLP
 
 | | E. coli Transcription Initiation Complex - 16-bp spacer and 5-nt RNA | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, Steitz, T.A. | | Deposit date: | 2015-03-05 | | Release date: | 2015-04-22 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Crystal Structures of the E. coli Transcription Initiation Complexes with a Complete Bubble.
Mol.Cell, 58, 2015
|
|
4YG2
 
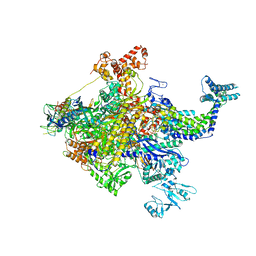 | | X-ray crystal structur of Escherichia coli RNA polymerase sigma70 holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Murakami, K.S. | | Deposit date: | 2015-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme
J. Biol. Chem., 288, 2013
|
|
